6 Flowline Distance
Purpose: calculate flowline distance between 6 mainstem Snake River sections (midpoints) and downstream extent of 52 reporting units
6.1 Load flowline
Load flowline as riverdist network
Code
flowline <- line2network(path = "/Users/jeffbaldock/Library/CloudStorage/GoogleDrive-jbaldock@uwyo.edu/Shared drives/wyo-coop-baldock/UWyoming/Snake River Cutthroat/Data/Spatial/Flowline/FlowlineEditing", layer = "SnakeHeadwaters_flowline_springsclean")
# plot(flowline)
# zoomtoseg(seg = c(67,3423), rivers = flowline)Fix topological errors (from raw flowline shp, mouth at segment 65 and vertex 94)
Load cleaned flowline object/riverdist network
Code
load(file = "/Users/jeffbaldock/Library/CloudStorage/GoogleDrive-jbaldock@uwyo.edu/Shared drives/wyo-coop-baldock/UWyoming/Snake River Cutthroat/Analyses/Snake River GSI Quarto/Landscape Covariates/Flowline Distance/SnakeHeadwaters_RiverDist_Cleaned.Rdata")6.2 Load points
Load reporting unit and mainstem sites and convert to shapefile
Code
# reporting units
sites_repu <- sf::read_sf(dsn = "/Users/jeffbaldock/Library/CloudStorage/GoogleDrive-jbaldock@uwyo.edu/Shared drives/wyo-coop-baldock/UWyoming/Snake River Cutthroat/Analyses/Snake River GSI Quarto/Landscape Covariates/Location Data", layer = "RepUnit_SpatialLocations")
# mainstem SR section midpoints
sites_mixt <- read_csv("/Users/jeffbaldock/Library/CloudStorage/GoogleDrive-jbaldock@uwyo.edu/Shared drives/wyo-coop-baldock/UWyoming/Snake River Cutthroat/Analyses/Snake River GSI Quarto/Landscape Covariates/Snake_section_midpoints.csv")
sites_mixt1 <- sf::st_as_sf(sites_mixt, coords = c("lon", "lat"), crs = 4326) %>% sf::st_transform(crs = sf::st_crs(sites_repu))
names(sites_mixt1) <- c("repunit", "geometry")Convert to river points and snap to network
Code
sites_repu2 <- xy2segvert(x = sf::st_coordinates(sites_repu)[,1], y = sf::st_coordinates(sites_repu)[,2], rivers = flowline_fixed)
sites_mixt2 <- xy2segvert(x = sf::st_coordinates(sites_mixt1)[,1], y = sf::st_coordinates(sites_mixt1)[,2], rivers = flowline_fixed)Inspect points. For example:
Code
zoomtoseg(seg = c(2077), rivers = flowline_fixed)
points(sf::st_coordinates(sites_repu)[,1], sf::st_coordinates(sites_repu)[,2], pch = 16, col = "red")
riverpoints(seg = sites_repu2$seg, vert = sites_repu2$vert, rivers = flowline_fixed, pch = 16, col = "blue")
6.3 Compute distances
Compute distances between reporting units and Snake River section mid-points
Code
distlist <- list()
for (i in 1:dim(sites_mixt1)[1]) {
sites <- rbind(sites_repu, sites_mixt1[i,])
sites2 <- xy2segvert(x = st_coordinates(sites)[,1], y = st_coordinates(sites)[,2], rivers = flowline_fixed)
disttib <- tibble(section = rep(sites_mixt1$repunit[i], times = 57), repunit = NA, distkm = NA)
for (j in 1:52) {
disttib$repunit[j] <- sites$repunit[j]
distm <- riverdistance(startseg = sites2$seg[53], startvert = sites2$vert[53],
endseg = sites2$seg[j], endvert = sites2$vert[j], rivers = flowline_fixed, map = F)
disttib$distkm[j] <- distm/1000
print(j)
}
distlist[[i]] <- disttib
}
disttib <- do.call(rbind, distlist)
write_csv(disttib, "Landscape Covariates/Flowline Distance/SnakeRiverSections_RepUnits_FlowlineDistance.csv")Load distances
Code
disttib <- read_csv("/Users/jeffbaldock/Library/CloudStorage/GoogleDrive-jbaldock@uwyo.edu/Shared drives/wyo-coop-baldock/UWyoming/Snake River Cutthroat/Analyses/Snake River GSI Quarto/Landscape Covariates/Flowline Distance/SnakeRiverSections_RepUnits_FlowlineDistance.csv")Plot distribution
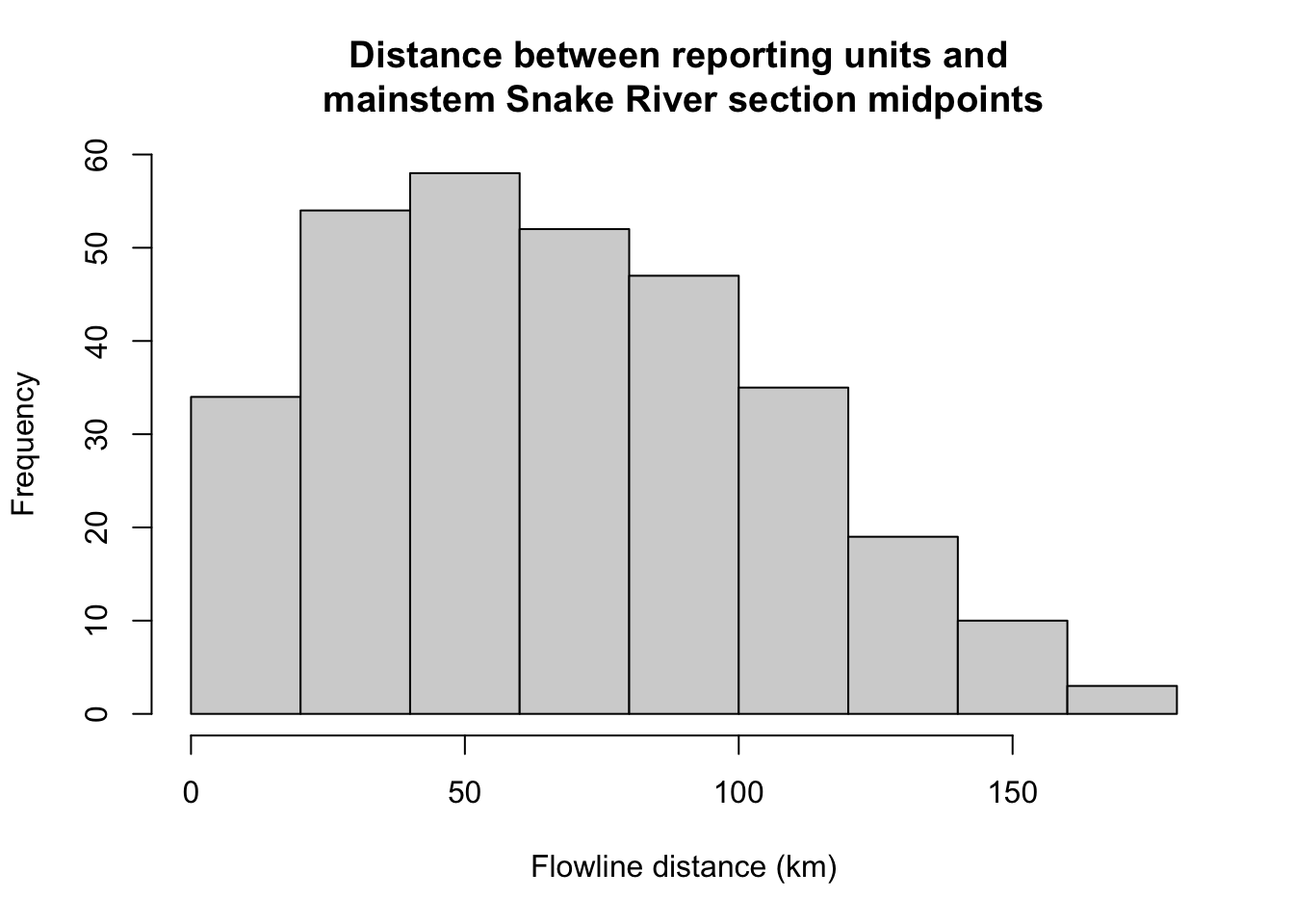
Code
hist(disttib$distkm, xlab = "Flowline distance (km)", main = "Distance between reporting units and \nmainstem Snake River section midpoints")