Code
siteinfo <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Data/EcoDrought_SiteInformation.csv")
siteinfo_sp <- st_as_sf(siteinfo, coords = c("long", "lat"), crs = 4326)Purpose: Quantify the suitability of existing modeling techniques (USGS StreamStats) for predicting streamflow in headwater systems.
Site information
siteinfo <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Data/EcoDrought_SiteInformation.csv")
siteinfo_sp <- st_as_sf(siteinfo, coords = c("long", "lat"), crs = 4326)Little g’s
dat_clean <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/LittleG_data_clean.csv")Big G’s
dat_clean_big <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/BigG_data_clean.csv")Climate
climdf <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/Daymet_climate.csv")
climdf_summ <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/Daymet_climate_summary.csv")Water availability
wateravail <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/BigG_wateravailability_annual.csv")Order sites: for colors, order sites from downstream to upstream (roughly) and by subbasin (if appropriate)
wborder <- c("West Brook NWIS", "West Brook Lower", "Mitchell Brook", "Jimmy Brook", "Obear Brook Lower", "West Brook Upper", "West Brook Reservoir", "Sanderson Brook", "Avery Brook", "West Whately Brook")
paineorder <- c("Paine Run 10", "Paine Run 08", "Paine Run 07", "Paine Run 06", "Paine Run 02", "Paine Run 01")
stauntorder <- c("Staunton River 10", "Staunton River 09", "Staunton River 07", "Staunton River 06", "Staunton River 03", "Staunton River 02")
flatorder <- c("BigCreekLower", "LangfordCreekLower", "LangfordCreekUpper", "Big Creek NWIS", "BigCreekUpper", "HallowattCreekLower", "NicolaCreek", "WernerCreek", "Hallowat Creek NWIS", "CoalCreekLower", "CycloneCreekLower", "CycloneCreekMiddle", "CycloneCreekUpper", "CoalCreekMiddle", "CoalCreekNorth", "CoalCreekHeadwaters", "McGeeCreekLower", "McGeeCreekTrib", "McGeeCreekUpper")
yellorder <- c("Shields River Valley Ranch", "Deep Creek", "Crandall Creek", "Buck Creek", "Dugout Creek", "Shields River ab Dugout", "Lodgepole Creek", "EF Duck Creek be HF", "EF Duck Creek ab HF", "Henrys Fork")
snakeorder <- c("Spread Creek Dam", "Rock Creek", "NF Spread Creek Lower", "NF Spread Creek Upper", "Grizzly Creek", "SF Spread Creek Lower", "Grouse Creek", "SF Spread Creek Upper", "Leidy Creek Mouth")
donnerorder <- c("Fish Creek NWIS", "Donner Blitzen ab Fish NWIS", "Donner Blitzen nr Burnt Car NWIS", "Donner Blitzen ab Indian NWIS")Write out point shape files for each state to feed into Stream Stats batch processor
siteinfo_sp_wy <- siteinfo_sp %>% filter(region == "Snake")
st_write(siteinfo_sp_wy, "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/points/points_wy.shp")
siteinfo_sp_mt <- siteinfo_sp %>% filter(region %in% c("Flat", "Shields"))
st_write(siteinfo_sp_mt, "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/points/points_mt.shp")
siteinfo_sp_ma <- siteinfo_sp %>% filter(region == "Mass")
st_write(siteinfo_sp_ma, "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/points/points_ma.shp")
siteinfo_sp_va <- siteinfo_sp %>% filter(region %in% c("Shen"))
st_write(siteinfo_sp_va, "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/points/points_va.shp")
siteinfo_sp_or <- siteinfo_sp %>% filter(region %in% c("Oreg"))
st_write(siteinfo_sp_or, "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/points/points_or.shp")List geodatabase layer names
st_layers(dsn = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/StreamStats/points_mt7617/points_mt7617.gdb")Driver: OpenFileGDB
Available layers:
layer_name geometry_type features fields crs_name
1 GlobalWatershedPoint Point 39 8 WGS 84
2 GlobalWatershed Multi Polygon 39 28 WGS 84
3 CHARACTERISTICS NA 1921 11 <NA>
4 FLOWSTATS NA 6763 16 <NA>Read watershed boundaries
sheds_montana <- st_read("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/StreamStats/points_mt7617/points_mt7617.gdb", layer = "GlobalWatershed")Reading layer `GlobalWatershed' from data source
`C:\Users\jbaldock\OneDrive - DOI\Documents\USGS\EcoDrought\EcoDrought Working\EcoDrought-Analysis\CompareModeledQ\StreamStats\points_mt7617\points_mt7617.gdb'
using driver `OpenFileGDB'
Simple feature collection with 39 features and 28 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -114.8904 ymin: 43.9457 xmax: -109.7226 ymax: 49.46148
Geodetic CRS: WGS 84sheds_massach <- st_read("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/StreamStats/points_ma7625/points_ma7625.gdb", layer = "GlobalWatershed")Reading layer `GlobalWatershed' from data source
`C:\Users\jbaldock\OneDrive - DOI\Documents\USGS\EcoDrought\EcoDrought Working\EcoDrought-Analysis\CompareModeledQ\StreamStats\points_ma7625\points_ma7625.gdb'
using driver `OpenFileGDB'
Simple feature collection with 13 features and 25 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -72.82306 ymin: 42.4123 xmax: -72.62871 ymax: 42.54973
Geodetic CRS: WGS 84sheds_oregon <- st_read("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/StreamStats/points_or7626/points_or7626.gdb", layer = "GlobalWatershed")Reading layer `GlobalWatershed' from data source
`C:\Users\jbaldock\OneDrive - DOI\Documents\USGS\EcoDrought\EcoDrought Working\EcoDrought-Analysis\CompareModeledQ\StreamStats\points_or7626\points_or7626.gdb'
using driver `OpenFileGDB'
Simple feature collection with 7 features and 41 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -118.9295 ymin: 42.48917 xmax: -118.561 ymax: 42.79204
Geodetic CRS: WGS 84sheds_virginia <- st_read("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/StreamStats/points_va7627/points_va7627.gdb", layer = "GlobalWatershed")Reading layer `GlobalWatershed' from data source
`C:\Users\jbaldock\OneDrive - DOI\Documents\USGS\EcoDrought\EcoDrought Working\EcoDrought-Analysis\CompareModeledQ\StreamStats\points_va7627\points_va7627.gdb'
using driver `OpenFileGDB'
Simple feature collection with 32 features and 20 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -79.24034 ymin: 37.88165 xmax: -78.02949 ymax: 38.7622
Geodetic CRS: WGS 84sheds_wyoming <- st_read("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/StreamStats/points_wy7628/points_wy7628.gdb", layer = "GlobalWatershed")Reading layer `GlobalWatershed' from data source
`C:\Users\jbaldock\OneDrive - DOI\Documents\USGS\EcoDrought\EcoDrought Working\EcoDrought-Analysis\CompareModeledQ\StreamStats\points_wy7628\points_wy7628.gdb'
using driver `OpenFileGDB'
Simple feature collection with 14 features and 17 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -110.5241 ymin: 43.64373 xmax: -110.1598 ymax: 43.87029
Geodetic CRS: WGS 84sheds <- bind_rows(sheds_massach, sheds_montana, sheds_oregon, sheds_virginia, sheds_wyoming)
mapview(sheds) Find sites that were delineated incorrectly
options(scipen=999)
badsites <- tibble(sheds) %>% select(Name, Shape_Area, DRNAREA, ELEV) %>% rename(site_id = Name) %>% left_join(siteinfo %>% select(site_id, site_name, area_sqmi, elev_ft)) %>% select(site_id, site_name, DRNAREA, area_sqmi) %>% mutate(percerror = (DRNAREA - area_sqmi) / area_sqmi) %>% filter(percerror >= 0.15 | percerror <= -0.15)
badsites# A tibble: 15 × 5
site_id site_name DRNAREA area_sqmi percerror
<chr> <chr> <dbl> <dbl> <dbl>
1 WW West Whately Brook 0.0399 0.493 -0.919
2 WL West Brook Lower 0.086 8.51 -0.990
3 JB Jimmy Brook 7.87 0.974 7.08
4 SH08 Shields River ab Dugout 11.1 8.68 0.279
5 SH06 Lodgepole Creek 2.2 1.36 0.619
6 SH05 Dugout Creek 11.1 2.39 3.64
7 BIG_002 LangfordCreekLower 0.1 3.99 -0.975
8 RAP Rapidan River NWIS 0.0000386 115 -1.000
9 PI_09FL Piney River 09 0.36 4.28 -0.916
10 LEI Leidy Creek Mouth NWIS 0.000811 5.17 -1.000
11 PCM Pacific Creek at Moran NWIS 0.34 166. -0.998
12 SP10 SF Spread Creek Upper 0.000348 35.1 -1.000
13 SP09 SF Spread Creek Lower 72 44.3 0.627
14 SP08 Rock Creek 0.0000772 4.74 -1.000
15 SP03 Leidy Creek Lower 0.00112 5.17 -1.000Stream stats site information
streamstats_info <- tibble(sheds) %>% select(Name, DRNAREA) %>% rename(site_id = Name) %>% left_join(siteinfo %>% select(site_id, site_name))
# streamstats_infoRead flow statistics from geodatabases
montana <- st_read("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/StreamStats/points_mt7617/points_mt7617.gdb", layer = "FLOWSTATS")Reading layer `FLOWSTATS' from data source
`C:\Users\jbaldock\OneDrive - DOI\Documents\USGS\EcoDrought\EcoDrought Working\EcoDrought-Analysis\CompareModeledQ\StreamStats\points_mt7617\points_mt7617.gdb'
using driver `OpenFileGDB'massach <- st_read("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/StreamStats/points_ma7625/points_ma7625.gdb", layer = "FLOWSTATS")Reading layer `FLOWSTATS' from data source
`C:\Users\jbaldock\OneDrive - DOI\Documents\USGS\EcoDrought\EcoDrought Working\EcoDrought-Analysis\CompareModeledQ\StreamStats\points_ma7625\points_ma7625.gdb'
using driver `OpenFileGDB'oregon <- st_read("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/StreamStats/points_or7626/points_or7626.gdb", layer = "FLOWSTATS")Reading layer `FLOWSTATS' from data source
`C:\Users\jbaldock\OneDrive - DOI\Documents\USGS\EcoDrought\EcoDrought Working\EcoDrought-Analysis\CompareModeledQ\StreamStats\points_or7626\points_or7626.gdb'
using driver `OpenFileGDB'virginia <- st_read("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/StreamStats/points_va7627/points_va7627.gdb", layer = "FLOWSTATS")Reading layer `FLOWSTATS' from data source
`C:\Users\jbaldock\OneDrive - DOI\Documents\USGS\EcoDrought\EcoDrought Working\EcoDrought-Analysis\CompareModeledQ\StreamStats\points_va7627\points_va7627.gdb'
using driver `OpenFileGDB'wyoming <- st_read("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/StreamStats/points_wy7628/points_wy7628.gdb", layer = "FLOWSTATS")Reading layer `FLOWSTATS' from data source
`C:\Users\jbaldock\OneDrive - DOI\Documents\USGS\EcoDrought\EcoDrought Working\EcoDrought-Analysis\CompareModeledQ\StreamStats\points_wy7628\points_wy7628.gdb'
using driver `OpenFileGDB'streamstats <- bind_rows(montana, massach, oregon, virginia, wyoming) %>% filter(!Name %in% c(badsites$site_id)) %>% rename(site_id = Name) %>% left_join(siteinfo %>% select(site_id, site_name)) %>% left_join(streamstats_info)
head(streamstats) site_id RegionID RegionName AreaPercent AreaSqMi
1 NFF GC1906 Crippen_Bue_Region_13 61 947.3549
2 NFF GC1828 USA_Bieger_2015 60 937.1283
3 NFF GC1828 USA_Bieger_2015 60 937.1283
4 NFF GC1828 USA_Bieger_2015 60 937.1283
5 NFF GC1818 Northern_Rocky_Mountains_P_Bieger_2015 60 937.1283
6 NFF GC1818 Northern_Rocky_Mountains_P_Bieger_2015 60 937.1283
StatLabel StatName Value
1 PKMAX_CB_R Maximum Flood Crippen Bue Regional 246000.00
2 XABNKF_U_B Bieger_USA_channel_cross_sectional_area 905.00
3 DBANKF_U_B Bieger_USA_channel_depth 5.77
4 WBANKF_U_B Bieger_USA_channel_width 165.00
5 XABNKF_P_B Bieger_P_channel_cross_sectional_area 852.00
6 DBANKF_P_B Bieger_P_channel_depth 5.53
Units Years PIl PIu SE SEp PC CitationID
1 cubic feet per second NA NA NA NA NA NA 186
2 square feet 0 NA NA NA NA NA 160
3 feet 0 NA NA NA NA NA 160
4 feet 0 NA NA NA NA NA 160
5 square feet 0 NA NA NA NA NA 160
6 feet 0 NA NA NA NA NA 160
site_name DRNAREA
1 North Fork Flathead River NWIS 1556.2
2 North Fork Flathead River NWIS 1556.2
3 North Fork Flathead River NWIS 1556.2
4 North Fork Flathead River NWIS 1556.2
5 North Fork Flathead River NWIS 1556.2
6 North Fork Flathead River NWIS 1556.2char_massach <- st_read("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/CompareModeledQ/StreamStats/points_ma7625/points_ma7625.gdb", layer = "CHARACTERISTICS")Reading layer `CHARACTERISTICS' from data source
`C:\Users\jbaldock\OneDrive - DOI\Documents\USGS\EcoDrought\EcoDrought Working\EcoDrought-Analysis\CompareModeledQ\StreamStats\points_ma7625\points_ma7625.gdb'
using driver `OpenFileGDB'head(char_massach) Name RegionID RegionName AreaPercent AreaSqMi StatLabel
1 WBR GC1894 Crippen_Bue_Region_1 100 10.70496 DRNAREA
2 WBR GC828 Perennial_Flow_Probability 100 10.70496 MAREGION
3 WBR GC828 Perennial_Flow_Probability 100 10.70496 FOREST
4 WBR GC828 Perennial_Flow_Probability 100 10.70496 PCTSNDGRV
5 WBR GC828 Perennial_Flow_Probability 100 10.70496 DRNAREA
6 WBR GC1828 USA_Bieger_2015 100 10.70496 DRNAREA
StatName Value
1 Area that drains to a point on a stream 10.7
2 Region of Massachusetts 0 for Eastern 1 for Western 1.0
3 Percentage of area covered by forest 87.7
4 Percentage of land surface underlain by sand and gravel deposits 14.7
5 Area that drains to a point on a stream 10.7
6 Area that drains to a point on a stream 10.7
Units MinLimit MaxLimit
1 square miles 0.10000 10000.00
2 dimensionless 0.00000 1.00
3 percent 0.00000 100.00
4 percent 0.00000 100.00
5 square miles 0.01000 1.99
6 square miles 0.07722 59927.74View provided flow statistics for each state. What is relevant to this paper?
print("MONTANA")
unique(montana$StatName)
print("MASSACHUSETTS")
unique(massach$StatName)
print("OREGON")
unique(oregon$StatName)
print("VIRGINA")
unique(virginia$StatName)
print("WYOMING")
unique(wyoming$StatName)Availability of flow statistics varies greatly by state. What statistics do all states have in common (minus Wyoming)? Only 7 day 10 year low flow is relevant.
Reduce(intersect, list(unique(montana$StatName), unique(massach$StatName), unique(oregon$StatName), unique(virginia$StatName))) [1] "Maximum Flood Crippen Bue Regional"
[2] "Bieger_USA_channel_cross_sectional_area"
[3] "Bieger_USA_channel_depth"
[4] "Bieger_USA_channel_width"
[5] "Bieger_P_channel_cross_sectional_area"
[6] "Bieger_P_channel_depth"
[7] "Bieger_P_channel_width"
[8] "Bieger_D_channel_cross_sectional_area"
[9] "Bieger_D_channel_depth"
[10] "Bieger_D_channel_width"
[11] "7 Day 10 Year Low Flow" For the West Brook, plot (annual) observed and StreamStats exceedance/duration curves and calculate absolute error
# set up
vars <- unique(massach$StatName)[grep("Duration", unique(massach$StatName))][-1]
sites <- c("West Brook NWIS", "Mitchell Brook", "Obear Brook Lower", "West Brook Upper", "Sanderson Brook", "Avery Brook")
preds <- list()
exceed <- list()
joined <- list()
joined_full <- list()
# calcualate
for (i in 1:length(sites)) {
obs <- dat_clean %>% filter(site_name == sites[i])
# stream stats duration
p <- streamstats %>%
filter(site_name == unique(obs$site_name), StatName %in% vars) %>%
mutate(exceedance = parse_number(StatName)) %>%
mutate(flow_cms = Value*0.02831683199881, area_sqkm = DRNAREA*2.58999)
p <- add_daily_yield(data = p %>% select(site_id, site_name, DRNAREA, area_sqkm, StatName, exceedance, flow_cms), values = flow_cms, basin_area = as.numeric(unique(p$area_sqkm)))
p <- p %>% mutate(logYield = log(Yield_mm))
preds[[i]] <- p
# calculate exceedance probability by site
exceeddat <- obs %>%
filter(!is.na(logYield)) %>%
arrange(desc(logYield)) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield))
exceed[[i]] <- exceeddat
# join observed and streamstats exceedance, calculate error
j <- exceeddat %>%
select(site_name, exceedance, logYield) %>%
mutate(exceedance = round(exceedance, digits = 0)) %>%
group_by(site_name, exceedance) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
left_join(p %>%
select(site_name, exceedance, logYield) %>%
rename(logYield_ss = logYield)) %>%
mutate(error_abs = logYield_ss - logYield,
error_abs_real = exp(logYield_ss) - exp(logYield),
error_rel = (exp(logYield_ss) - exp(logYield)) / exp(logYield))
joined[[i]] <- j %>% filter(!is.na(error_abs))
joined_full[[i]] <- j
}
preds <- do.call(rbind, preds) %>% mutate(site_name = factor(site_name, levels = wborder))
exceed <- do.call(rbind, exceed) %>% mutate(site_name = factor(site_name, levels = wborder))
joined <- do.call(rbind, joined) %>% mutate(site_name = factor(site_name, levels = wborder))
joined_full <- do.call(rbind, joined_full)
joined_mean <- joined %>% group_by(exceedance) %>% summarize(error_abs = mean(error_abs, na.rm = TRUE))
# preds <- preds %>% mutate(site_name = factor(site_name, levels = sites))
# exceed <- exceed %>% mutate(site_name = factor(site_name, levels = sites))
# joined <- joined %>% mutate(site_name = factor(site_name, levels = sites))
# calculate among size variation for StreamStats and observed exceedance
vardat_ss <- joined %>% group_by(exceedance) %>% summarize(exdsd = sd(logYield_ss))
vardat_obs <- joined_full %>% group_by(exceedance) %>% summarize(exdsd = sd(logYield))
# tibble for site labels
siteslabs <- tibble(site_name = factor(sites, levels = sites), site_lab = c("WB0", "Mitchell", "Obear Lower", "WB Upper", "Sanderson", "Avery"))Plot observed (colored lines) and modeled (black points/lines) exceedance curves
mypal <- cet_pal(length(wborder), name = "i1")[c(1,3,5,6,8,9)]
### Colored by site
# exceedance curves
p1 <- ggplot() +
geom_line(data = exceed, aes(x = exceedance, y = logYield, color = site_name), size = 1) +
geom_line(data = preds, aes(x = exceedance, y = logYield), color = "black") +
geom_point(data = preds, aes(x = exceedance, y = logYield), color = "black") +
geom_text(data = siteslabs, aes(x = Inf, y = Inf, label = site_lab), vjust = 1.5, hjust = 1.05, size = 3) +
facet_wrap(~site_name, nrow = 3) +
scale_color_manual(values = mypal) +
xlab("Exceedance probability") + ylab(expression(paste("log(specific discharge, mm day"^-1, ")", sep = ""))) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.grid = element_blank(), strip.text.x = element_blank(), strip.background = element_blank(),
legend.position = "none", axis.text = element_text(color = "black"))
# absolute error
p2 <- ggplot(data = joined) +
geom_line(aes(x = exceedance, y = error_abs, group = site_name, color = site_name)) +
geom_point(aes(x = exceedance, y = error_abs, group = site_name, color = site_name)) +
geom_abline(intercept = 0, slope = 0, linetype = "dashed") +
geom_line(data = joined_mean, aes(x = exceedance, y = error_abs), size = 1) +
scale_color_manual(values = mypal) +
xlab("Exceedance probability") + ylab("Error (modeled - observed)") + ylim(-2.1,0) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
legend.position = "none", axis.text = element_text(color = "black"))
# combine
annotate_figure(ggarrange(p1, p2, ncol = 2, labels = "auto"), top = text_grob("The West Brook, Massachusetts"))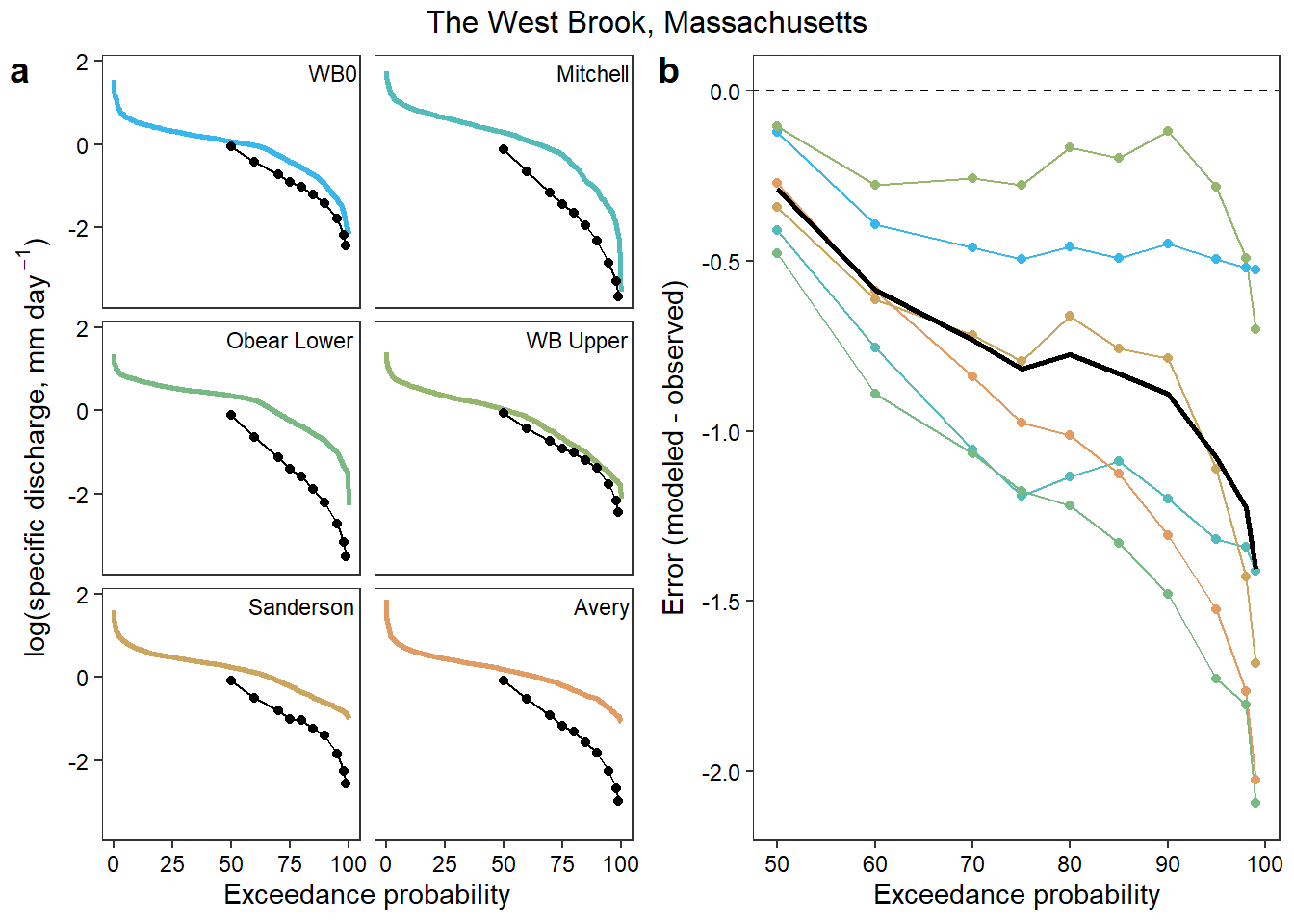
Does StreamStats misrepresent among-site variation in flow duration?
p1 <- ggplot() +
geom_line(data = vardat_obs, aes(x = exceedance, y = exdsd), size = 1, color = "grey60") +
geom_line(data = vardat_ss, aes(x = exceedance, y = exdsd), color = "black") +
geom_point(data = vardat_ss, aes(x = exceedance, y = exdsd), color = "black") +
xlab("Exceedance probability") + ylab("Among-site standard deviation in log(Yield, mm)") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
p2 <- vardat_obs %>%
left_join(vardat_ss %>% rename(exdsd_ss = exdsd)) %>%
mutate(diff = exdsd_ss - exdsd) %>%
filter(!is.na(diff)) %>%
ggplot() +
geom_line(aes(x = exceedance, y = diff)) +
geom_abline(intercept = 0, slope = 0, linetype = "dashed") +
xlab("Exceedance probability") + ylab("Difference") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
annotate_figure(ggarrange(p1, p2, ncol = 2, labels = "auto"), top = text_grob("The West Brook, Massachusetts"))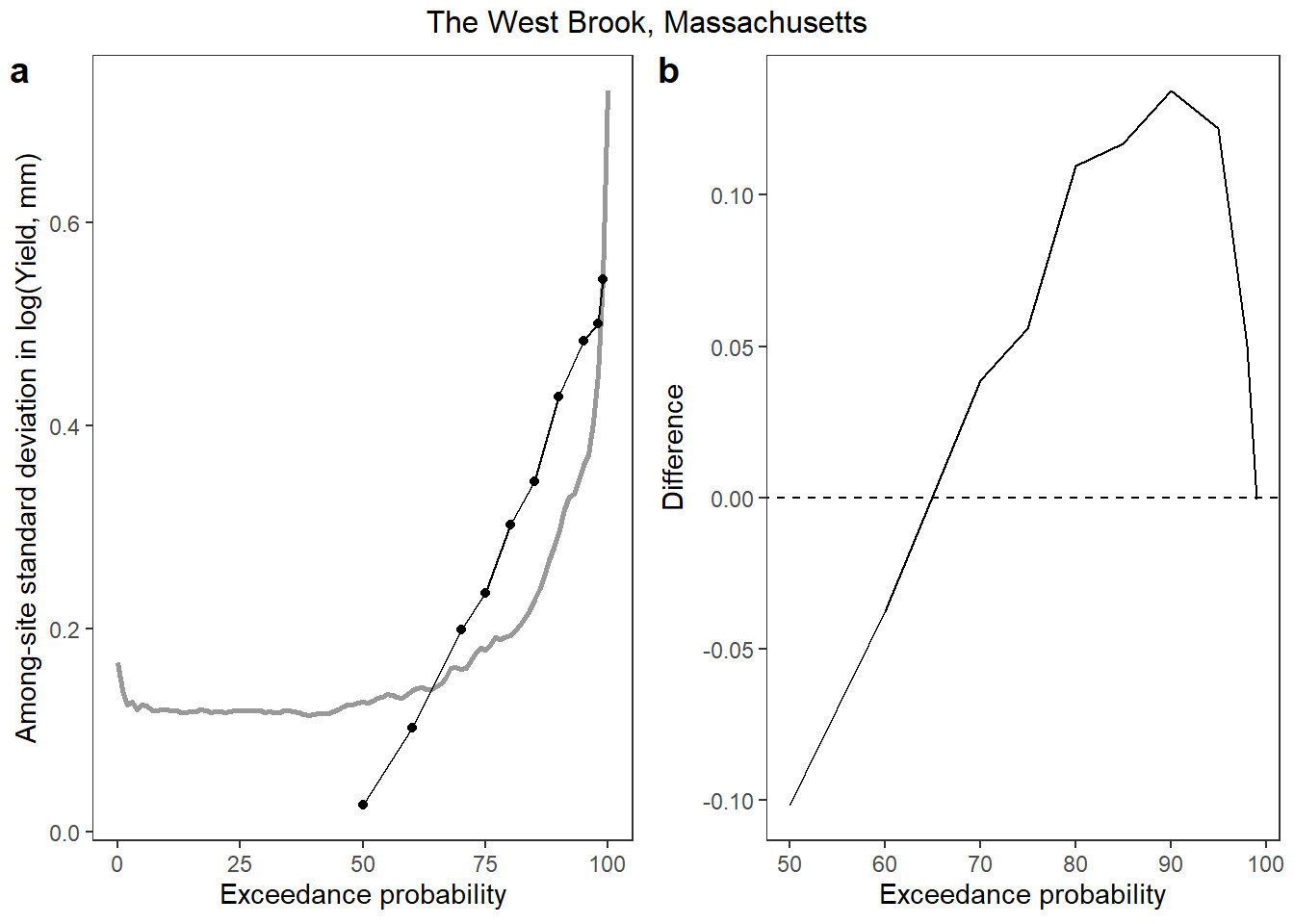
Write to file
mypal <- cet_pal(length(wborder), name = "i1")[c(1,3,5,6,8,9)]
### Colored by site
# exceedance curves
p1 <- ggplot() +
geom_line(data = exceed, aes(x = exceedance, y = logYield, color = site_name), size = 1) +
geom_line(data = preds, aes(x = exceedance, y = logYield), color = "black") +
geom_point(data = preds, aes(x = exceedance, y = logYield), color = "black") +
geom_text(data = siteslabs, aes(x = Inf, y = Inf, label = site_lab), vjust = 1.5, hjust = 1.05, size = 3) +
facet_wrap(~site_name, nrow = 3) +
scale_color_manual(values = mypal) +
xlab("Exceedance probability") + ylab(expression(paste("log(specific discharge, mm day"^-1, ")", sep = ""))) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.grid = element_blank(), strip.text.x = element_blank(), strip.background = element_blank(),
legend.position = "none", axis.text = element_text(color = "black"))
# absolute error
p2 <- ggplot(data = joined) +
geom_line(aes(x = exceedance, y = error_abs, group = site_name, color = site_name)) +
geom_point(aes(x = exceedance, y = error_abs, group = site_name, color = site_name)) +
geom_abline(intercept = 0, slope = 0, linetype = "dashed") +
geom_line(data = joined_mean, aes(x = exceedance, y = error_abs), size = 1) +
scale_color_manual(values = mypal) +
xlab("Exceedance probability") + ylab("Error (modeled - observed)") + ylim(-2.1,0) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
legend.position = "none", axis.text = element_text(color = "black"))
# combine
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/StreamStats_WestBrook.jpg", width = 6, height = 4, units = "in", res = 1000)
annotate_figure(ggarrange(p1, p2, ncol = 2, labels = "auto"), top = text_grob("The West Brook, Massachusetts"))
dev.off()For the Flathead (Big Creek), plot observed and StreamStats mean monthly flow/yield and calculate absolute error
# set up
vars <- unique(montana$StatName)[grep("Mean Flow", unique(montana$StatName))]
sites <- c("LangfordCreekUpper", "Big Creek NWIS", "HallowattCreekLower", "WernerCreek", "Hallowat Creek NWIS", "McGeeCreekLower")
preds_list <- list()
obs_list <- list()
hull_list <- list()
join_list <- list()
# calcualate
for (i in 1:length(sites)) {
# filter observed data
obs <- dat_clean %>%
filter(site_name == sites[i]) %>%
mutate(MonthName = factor(MonthName, levels = c("Oct", "Nov", "Dec", "Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sep")))
# calculate monthly means
obs_mon <- obs %>%
group_by(site_name, WaterYear, MonthName) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
mutate(MonthName = factor(MonthName, levels = c("Oct", "Nov", "Dec", "Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sep")),
WaterYear = factor(WaterYear)) %>%
complete(site_name, WaterYear, MonthName)
# get monthly min/max for ribbon
hull <- obs_mon %>%
group_by(site_name, MonthName) %>%
summarize(min_logYield = min(logYield, na.rm = TRUE), max_logYield = max(logYield, na.rm = TRUE)) %>%
ungroup()
# get StreamStats mean monthly flow
preds <- streamstats %>%
filter(site_name == sites[i], StatName %in% vars, !is.na(AreaSqMi)) %>%
mutate(MonthName = substr(StatName, 1, nchar(StatName)-10),
Month = parse_number(StatLabel)) %>%
mutate(MonthName = factor(recode(MonthName, "January" = "Jan", "February" = "Feb", "March" = "Mar", "April" = "Apr", "June" = "Jun", "July" = "Jul", "August" = "Aug", "September" = "Sep", "October" = "Oct", "November" = "Nov", "December" = "Dec"), levels = c("Oct", "Nov", "Dec", "Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sep"))) %>%
mutate(flow_cms = Value*0.02831683199881, area_sqkm = AreaSqMi*2.58999)
preds <- add_daily_yield(data = preds %>% select(site_id, site_name, area_sqkm, MonthName, Month, flow_cms), values = flow_cms, basin_area = as.numeric(unique(preds$area_sqkm)))
preds <- preds %>% mutate(logYield = log(Yield_mm))
# join and calculate error
join_list[[i]] <- obs_mon %>%
left_join(preds %>% select(site_name, MonthName, logYield) %>% rename(logYield_ss = logYield)) %>%
mutate(error_abs = logYield_ss - logYield,
error_abs_real = exp(logYield_ss) - exp(logYield),
error_rel = (exp(logYield_ss) - exp(logYield)) / exp(logYield))
# store in list
preds_list[[i]] <- preds
obs_list[[i]] <- obs_mon
hull_list[[i]] <- hull
}
preds <- do.call(rbind, preds_list) %>% mutate(site_name = factor(site_name, levels = flatorder))
obs_mon <- do.call(rbind, obs_list) %>% mutate(site_name = factor(site_name, levels = flatorder))
hull <- do.call(rbind, hull_list) %>% mutate(site_name = factor(site_name, levels = flatorder))
joined <- do.call(rbind, join_list) %>% mutate(site_name = factor(site_name, levels = flatorder))
joined_mean <- joined %>% group_by(MonthName) %>% summarise(error_abs = mean(error_abs, na.rm = TRUE))
# tibble for site labels
siteslabs <- tibble(site_name = factor(sites, levels = flatorder), site_lab = c("Langford", "Big", "Hallowatt Lower", "Werner", "Hallowatt Upper", "McGee"))
# calculate among site variation for StreamStats and observed mean monthly flow (mean across years)
vardat <- preds %>%
group_by(MonthName) %>%
summarize(qsd_ss = sd(logYield)) %>%
ungroup() %>%
left_join(obs_mon %>%
group_by(site_name, MonthName) %>%
summarize(logYield = mean(logYield, na.rm = TRUE)) %>%
ungroup() %>%
group_by(MonthName) %>%
summarize(qsd_obs = sd(logYield)) %>%
ungroup()) %>%
mutate(diff = qsd_ss - qsd_obs, nummon = as.numeric(MonthName))Plot observed and modeled mean monthly flow
mypal <- cet_pal(length(flatorder), name = "i1")[c(3,4,6,8,9,17)]
### Colored by site
# observed and StreamStats monthly flow
p1 <- ggplot() +
geom_ribbon(data = hull, aes(ymin = min_logYield, ymax = max_logYield, x = as.numeric(MonthName), fill = site_name), alpha = 0.3) +
geom_line(data = obs_mon, aes(y = logYield, x = as.numeric(MonthName), group = WaterYear, color = site_name)) +
geom_point(data = obs_mon, aes(y = logYield, x = as.numeric(MonthName), group = WaterYear, shape = WaterYear, color = site_name)) +
geom_line(data = preds, aes(y = logYield, x = as.numeric(MonthName), group = site_name), color = "black") +
geom_point(data = preds, aes(y = logYield, x = as.numeric(MonthName)), color = "black") +
geom_text(data = siteslabs, aes(x = -Inf, y = Inf, label = site_lab), vjust = 1.5, hjust = -0.05, size = 3) +
facet_wrap(~site_name, ncol = 2) +
scale_color_manual(values = mypal) +
scale_fill_manual(values = mypal) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
strip.text.x = element_blank(), strip.background = element_blank()) +
ylab(expression(paste("Monthly mean log(specific discharge, mm day"^-1, ")", sep = ""))) + xlab("Month") +
scale_x_continuous(breaks = 1:12, labels = c("O", "N", "D", "J", "F", "M", "A", "M", "J", "J", "A", "S"))
# absolute error by month
p2 <- ggplot() +
geom_line(data = joined, aes(x = as.numeric(MonthName), y = error_abs, shape = WaterYear, color = site_name)) +
geom_point(data = joined, aes(x = as.numeric(MonthName), y = error_abs, shape = WaterYear, color = site_name)) +
geom_line(data = joined_mean, aes(x = as.numeric(MonthName), y = error_abs), size = 1) +
geom_abline(intercept = 0, slope = 0, linetype = "dashed") +
scale_color_manual(values = mypal) +
xlab("Month") + ylab("Error (modeled - observed)") +
scale_x_continuous(breaks = 1:12, labels = c("O", "N", "D", "J", "F", "M", "A", "M", "J", "J", "A", "S")) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
legend.position = "none")
# combine
annotate_figure(ggarrange(p1, p2, ncol = 2, labels = "auto"), top = text_grob("North Fork Flathead River, Montana"))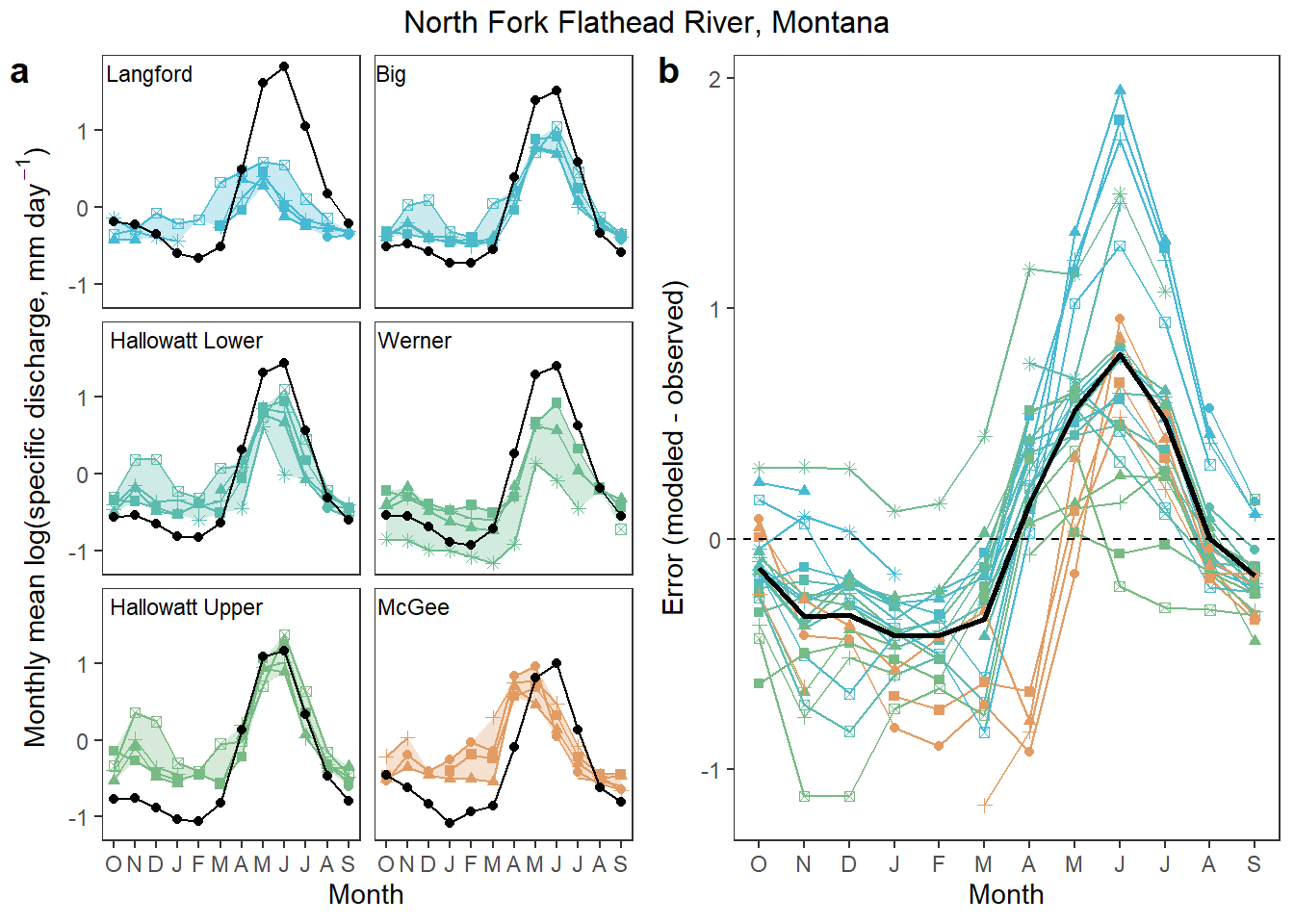
Does StreamStats misrepresent among-site variation in mean monthly yield?
p1 <- vardat %>%
ggplot() +
geom_line(aes(x = nummon, y = qsd_obs), size = 1, color = "grey60") +
geom_line(aes(x = nummon, y = qsd_ss), color = "black") +
geom_point(aes(x = nummon, y = qsd_ss), color = "black") +
xlab("Month") + ylab("Among-site standard deviation in log(Yield, mm)") +
scale_x_continuous(breaks = 1:12, labels = c("O", "N", "D", "J", "F", "M", "A", "M", "J", "J", "A", "S")) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
p2 <- vardat %>%
ggplot() +
geom_line(aes(x = nummon, y = diff)) +
xlab("Month") + ylab("Difference") +
geom_abline(intercept = 0, slope = 0, linetype = "dashed") +
scale_x_continuous(breaks = 1:12, labels = c("O", "N", "D", "J", "F", "M", "A", "M", "J", "J", "A", "S")) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
annotate_figure(ggarrange(p1, p2, ncol = 2, labels = "auto"), top = text_grob("North Fork Flathead River, Montana"))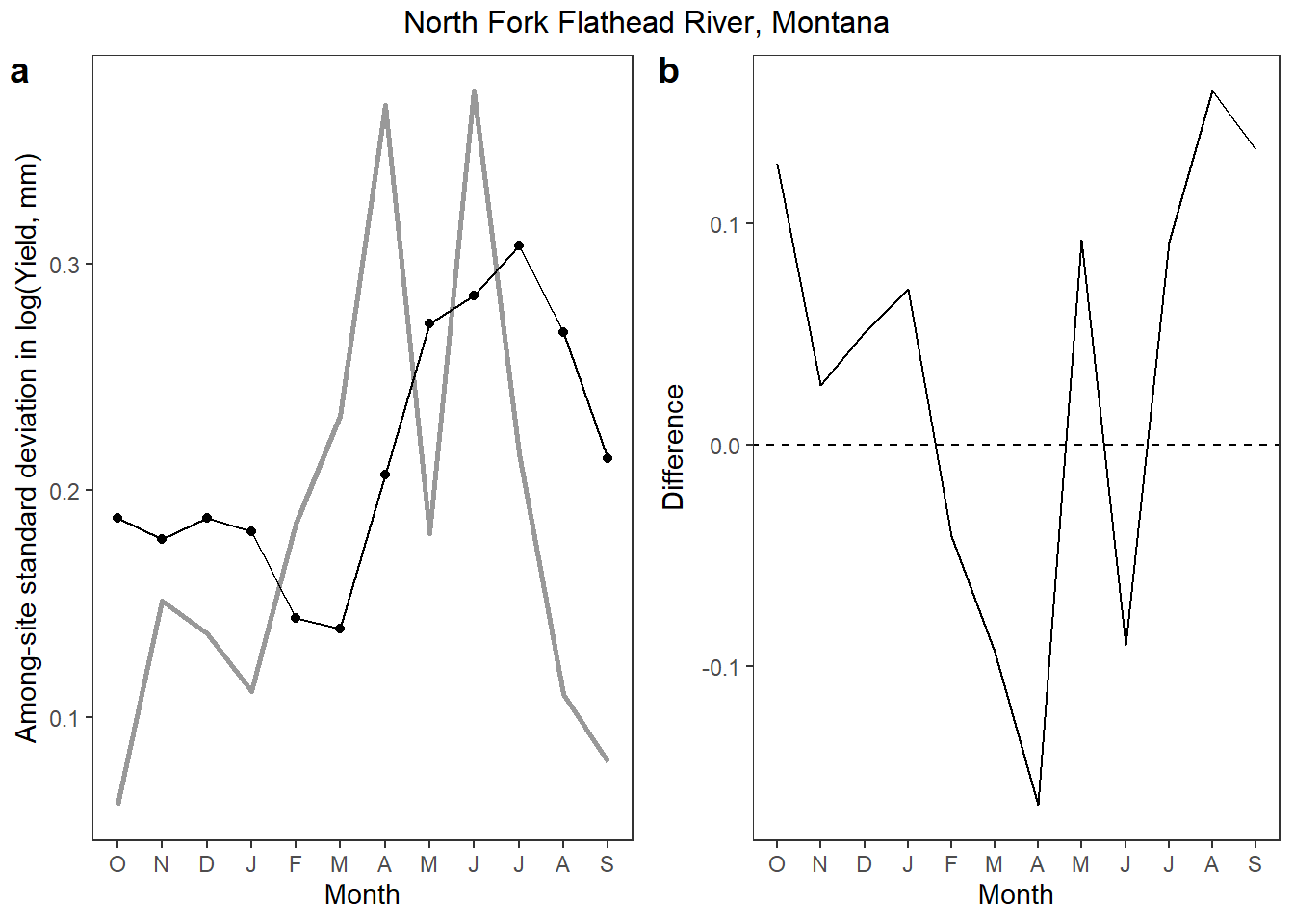
Write to file
mypal <- cet_pal(length(flatorder), name = "i1")[c(3,4,6,8,9,17)]
### Colored by site
# observed and StreamStats monthly flow
p1 <- ggplot() +
geom_ribbon(data = hull, aes(ymin = min_logYield, ymax = max_logYield, x = as.numeric(MonthName), fill = site_name), alpha = 0.3) +
geom_line(data = obs_mon, aes(y = logYield, x = as.numeric(MonthName), group = WaterYear, color = site_name)) +
geom_point(data = obs_mon, aes(y = logYield, x = as.numeric(MonthName), group = WaterYear, shape = WaterYear, color = site_name)) +
geom_line(data = preds, aes(y = logYield, x = as.numeric(MonthName), group = site_name), color = "black") +
geom_point(data = preds, aes(y = logYield, x = as.numeric(MonthName)), color = "black") +
geom_text(data = siteslabs, aes(x = -Inf, y = Inf, label = site_lab), vjust = 1.5, hjust = -0.05, size = 3) +
facet_wrap(~site_name, ncol = 2) +
scale_color_manual(values = mypal) +
scale_fill_manual(values = mypal) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
strip.text.x = element_blank(), strip.background = element_blank()) +
ylab(expression(paste("Monthly mean log(specific discharge, mm day"^-1, ")", sep = ""))) + xlab("Month") +
scale_x_continuous(breaks = 1:12, labels = c("O", "N", "D", "J", "F", "M", "A", "M", "J", "J", "A", "S"))
# absolute error by month
p2 <- ggplot() +
geom_line(data = joined, aes(x = as.numeric(MonthName), y = error_abs, shape = WaterYear, color = site_name)) +
geom_point(data = joined, aes(x = as.numeric(MonthName), y = error_abs, shape = WaterYear, color = site_name)) +
geom_line(data = joined_mean, aes(x = as.numeric(MonthName), y = error_abs), size = 1) +
geom_abline(intercept = 0, slope = 0, linetype = "dashed") +
scale_color_manual(values = mypal) +
xlab("Month") + ylab("Error (modeled - observed)") +
scale_x_continuous(breaks = 1:12, labels = c("O", "N", "D", "J", "F", "M", "A", "M", "J", "J", "A", "S")) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
legend.position = "none")
# combine
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/StreamStats_Flathead.jpg", width = 6, height = 4, units = "in", res = 1000)
annotate_figure(ggarrange(p1, p2, ncol = 2), top = text_grob("North Fork Flathead River, Montana"))
dev.off()