Code
siteinfo <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Data/EcoDrought_SiteInformation.csv")
siteinfo_sp <- st_as_sf(siteinfo, coords = c("long", "lat"), crs = 4326)Purpose: Evaluate the extent and magnitude of spatial and temporal variation in headwater streamflow regimes.
Site information
siteinfo <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Data/EcoDrought_SiteInformation.csv")
siteinfo_sp <- st_as_sf(siteinfo, coords = c("long", "lat"), crs = 4326)Little g’s (headwater sites)
dat_clean <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/LittleG_data_clean.csv")Big G’s (reference gages)
dat_clean_big <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/BigG_data_clean.csv")Climate
climdf <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/Daymet_climate.csv")
climdf_summ <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/Daymet_climate_summary.csv")Get catchment shapefiles
sheds_list <- list()
myfiles <- list.files(path = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/", pattern = ".shp")
for (i in 1:length(myfiles)) {
sheds_list[[i]] <- st_read(paste("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/", myfiles[i], sep = ""))
}
sheds <- do.call(rbind, sheds_list) %>%
mutate(site_id = ifelse(site_id == "SP01", "SP07", ifelse(site_id == "SP07", "SP01", site_id))) %>%
left_join(siteinfo)
#mapview(sheds %>% arrange(desc(area_sqmi)), alpha.regions = 0.2)For colors, order sites from downstream to upstream (roughly) and by subbasin (if appropriate)
wborder <- c("West Brook NWIS", "West Brook Lower", "Mitchell Brook", "Jimmy Brook", "Obear Brook Lower", "West Brook Upper", "West Brook Reservoir", "Sanderson Brook", "Avery Brook", "West Whately Brook")
paineorder <- c("Paine Run 10", "Paine Run 08", "Paine Run 07", "Paine Run 06", "Paine Run 02", "Paine Run 01")
stauntorder <- c("Staunton River 10", "Staunton River 09", "Staunton River 07", "Staunton River 06", "Staunton River 03", "Staunton River 02")
flatorder <- c("BigCreekLower", "LangfordCreekLower", "LangfordCreekUpper", "Big Creek NWIS", "BigCreekUpper", "HallowattCreekLower", "NicolaCreek", "WernerCreek", "Hallowat Creek NWIS", "CoalCreekLower", "CycloneCreekLower", "CycloneCreekMiddle", "CycloneCreekUpper", "CoalCreekMiddle", "CoalCreekNorth", "CoalCreekHeadwaters", "McGeeCreekLower", "McGeeCreekTrib", "McGeeCreekUpper")
yellorder <- c("Shields River Valley Ranch", "Deep Creek", "Crandall Creek", "Buck Creek", "Dugout Creek", "Shields River ab Dugout", "Lodgepole Creek", "EF Duck Creek be HF", "EF Duck Creek ab HF", "Henrys Fork")
snakeorder <- c("Spread Creek Dam", "Rock Creek", "NF Spread Creek Lower", "NF Spread Creek Upper", "Grizzly Creek", "SF Spread Creek Lower", "Grouse Creek", "SF Spread Creek Upper", "Leidy Creek Mouth")
donnerorder <- c("Fish Creek NWIS", "Donner Blitzen ab Fish NWIS", "Donner Blitzen nr Burnt Car NWIS", "Donner Blitzen ab Indian NWIS")View daily time series data by sub-basin. Note that we are using the “Super G” NWIS data for the reference gage (black line). Per Robert comment, entirely nested design is cute, but doesn’t reflect how the data is actually used.
Big G NWIS sites/reference gages for each basin/subbasin:
dat_clean_big %>% group_by(region, basin, subbasin) %>% summarize(site_name = unique(site_name)) %>% ungroup() %>% filter(!is.na(region)) %>% kable()| region | basin | subbasin | site_name |
|---|---|---|---|
| Flat | Flathead | Flathead | North Fork Flathead River NWIS |
| Mass | West Brook | West Brook | South River Conway NWIS |
| Oreg | Donner Blitzen | Donner Blitzen | Donner Blitzen River nr Frenchglen NWIS |
| Shen | Paine Run | Paine Run | South River Harriston NWIS |
| Shen | Piney River | Piney River | Battle Run NWIS |
| Shen | Staunton River | Staunton River | Rapidan River NWIS |
| Shields | Shields River | Shields River | Yellowstone River Livingston NWIS |
| Snake | Snake River | Snake River | Pacific Creek at Moran NWIS |
dat_clean %>%
filter(basin == "Flathead") %>%
bind_rows(dat_clean_big %>% filter(basin == "Flathead", date >= date("2017-07-29"), date <= date("2023-12-11"))) %>%
select(date, site_name, logYield) %>%
spread(key = site_name, value = logYield) %>%
relocate("North Fork Flathead River NWIS", .after = last_col()) %>%
dygraph() %>% dyRangeSelector() %>% dyAxis("y", label = "ln(Yield, mm)") %>%
#dyOptions(colors = c(brewer.pal(7, "Dark2"), "black")) %>%
dySeries("North Fork Flathead River NWIS", strokeBorderWidth = 1, strokeWidth = 1.5, color = "black")dat_clean %>%
filter(basin == "Shields River") %>%
bind_rows(dat_clean_big %>% filter(basin == "Shields River", date >= date("2015-04-01"), date <= date("2023-12-31"))) %>%
select(date, site_name, logYield) %>%
spread(key = site_name, value = logYield) %>%
relocate("Yellowstone River Livingston NWIS", .after = last_col()) %>%
dygraph() %>% dyRangeSelector() %>% dyAxis("y", label = "ln(Yield, mm)") %>%
#dyOptions(colors = c(brewer.pal(7, "Dark2"), "black")) %>%
dySeries("Yellowstone River Livingston NWIS", strokeBorderWidth = 1, strokeWidth = 1.5, color = "black")Create time series data plots objects
polycol <- "grey90"
mycols <- brewer.pal(7, "Set2")
### WEST BROOK
tempdat <- dat_clean %>%
filter(basin == "West Brook") %>%
mutate(site_name = factor(site_name, levels = wborder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "West Brook", date >= date("2020-01-01"), date <= date("2025-01-03"))
# color
pwb1 <- ggplot() +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2023-07-01"), xmax = ymd("2023-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2024-07-01"), xmax = ymd("2024-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = wborder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
ylim(-3,2) +
with_outer_glow(annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "West Brook, MA", hjust = 0, vjust = 1), colour = "white", expand = 10) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[1], fill = NA, linewidth = 1.5)
)
# greyscale
pwb2 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, group = site_name), color = "grey") +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "black", size = 1) +
ylim(-3,2) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "West Brook, MA", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### STAUNTON RIVER
tempdat <- dat_clean %>%
filter(basin == "Staunton River") %>%
mutate(site_name = factor(site_name, levels = stauntorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Staunton River", date >= as_date("2018-11-07"), date <= as_date("2022-10-19"))
# color
pst1 <- ggplot() +
geom_rect(aes(xmin = ymd("2019-07-01"), xmax = ymd("2019-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = stauntorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
with_outer_glow(annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Staunton River, VA", hjust = 0, vjust = 1), colour = "white", expand = 10) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[2], fill = NA, linewidth = 1.5)
)
# greyscale
pst2 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, group = site_name), color = "grey") +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "black", size = 1) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Staunton River", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black"), panel.border = element_rect(colour = mycols[2], fill = NA, linewidth = 2))
### PAINE RUN
tempdat <- dat_clean %>%
filter(basin == "Paine Run") %>%
mutate(site_name = factor(site_name, levels = paineorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Paine Run", date >= as_date("2018-11-07"), date <= as_date("2023-05-15"))
# color
ppa1 <- ggplot() +
geom_rect(aes(xmin = ymd("2019-07-01"), xmax = ymd("2019-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = paineorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
with_outer_glow(annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Paine Run, VA", hjust = 0, vjust = 1), colour = "white", expand = 10) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[3], fill = NA, linewidth = 1.5)
)
# greyscale
ppa2 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, group = site_name), color = "grey") +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "black", size = 1) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Paine Run", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### FLATHEAD
tempdat <- dat_clean %>%
filter(basin == "Flathead", date >= date("2018-07-29"), date <= date("2023-08-11")) %>%
mutate(site_name = factor(site_name, levels = flatorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Flathead", date >= date("2018-07-29"), date <= date("2023-08-11"))
# color
pfl1 <- ggplot() +
geom_rect(aes(xmin = ymd("2018-07-29"), xmax = ymd("2018-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2019-07-01"), xmax = ymd("2019-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2023-07-01"), xmax = ymd("2023-08-11"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = flatorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
with_outer_glow(annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Flathead River, MT", hjust = 0, vjust = 1), colour = "white", expand = 10) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[4], fill = NA, linewidth = 1.5)
)
# greyscale
pfl2 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, group = site_name), color = "grey") +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "black", size = 1) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Flathead River", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### YELLOWSTONE
tempdat <- dat_clean %>%
filter(basin == "Shields River", date >= date("2016-06-01"), date <= date("2023-11-01")) %>%
mutate(site_name = factor(site_name, levels = yellorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Shields River", date >= date("2016-06-01"), date <= date("2023-11-01"))
# color
pye1 <- ggplot() +
geom_rect(aes(xmin = ymd("2016-07-01"), xmax = ymd("2016-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2017-07-01"), xmax = ymd("2017-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2018-07-01"), xmax = ymd("2018-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2019-07-01"), xmax = ymd("2019-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2023-07-01"), xmax = ymd("2023-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = yellorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
with_outer_glow(annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Yellowstone River, MT", hjust = 0, vjust = 1), colour = "white", expand = 10) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[5], fill = NA, linewidth = 1.5)
)
# greyscale
pye2 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, group = site_name), color = "grey") +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "black", size = 1) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Yellowstone River, MT", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### SNAKE
tempdat <- dat_clean %>%
filter(basin == "Snake River", date >= date("2016-06-01"), date <= date("2023-10-03")) %>%
mutate(site_name = factor(site_name, levels = snakeorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Snake River", date >= date("2016-06-01"), date <= date("2023-10-03"))
# color
psn1 <- ggplot() +
geom_rect(aes(xmin = ymd("2016-07-01"), xmax = ymd("2016-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2017-07-01"), xmax = ymd("2017-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2018-07-01"), xmax = ymd("2018-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2019-07-01"), xmax = ymd("2019-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2023-07-01"), xmax = ymd("2023-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = snakeorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
with_outer_glow(annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Snake River, WY", hjust = 0, vjust = 1), colour = "white", expand = 10) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[6], fill = NA, linewidth = 1.5)
)
# greyscale
psn2 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, group = site_name), color = "grey") +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "black", size = 1) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Snake River, WY", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### DONNER BLITZEN
tempdat <- dat_clean %>%
filter(basin == "Donner Blitzen") %>%
mutate(site_name = factor(site_name, levels = donnerorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Donner Blitzen", date >= date("2019-07-15"), date <= as_date("2022-10-30"))
# color
pdb1 <- ggplot() +
geom_rect(aes(xmin = ymd("2019-07-15"), xmax = ymd("2019-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = donnerorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
with_outer_glow(annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Donner und Blitzen River, OR", hjust = 0, vjust = 1), colour = "white", expand = 10) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[7], fill = NA, linewidth = 1.5)
)
# greyscale
pdb2 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, group = site_name), color = "grey") +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "black", size = 1) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Donner und Blitzen River", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))Generate combined plot
annotate_figure(egg::ggarrange(pwb1, pst1, ppa1, pfl1, pye1, psn1, pdb1, ncol = 1), left = "log(Yield, mm/day)")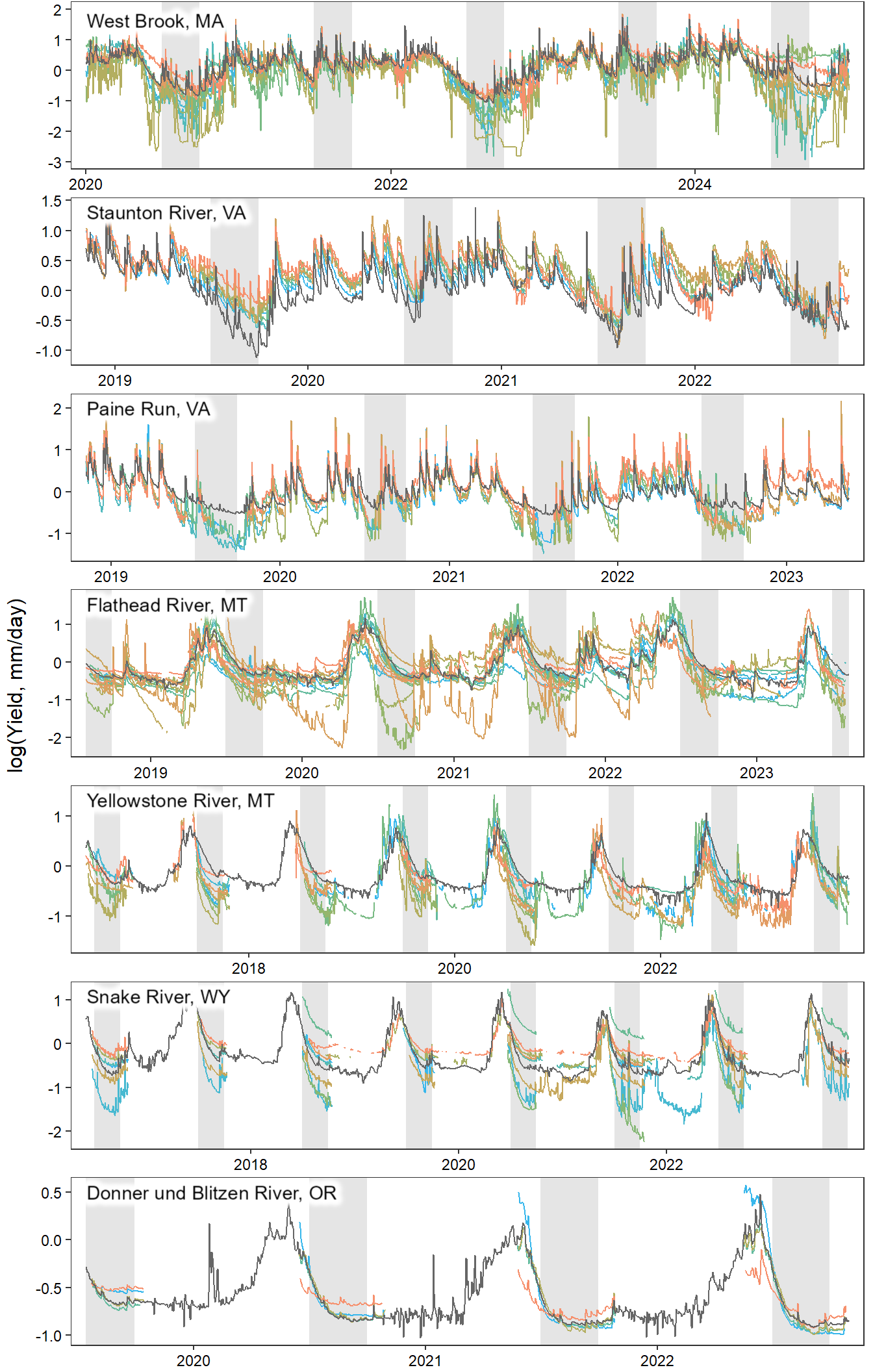
Write to file
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_timeseries_new_noborder.jpg", width = 5, height = 11, units = "in", res = 1000)
annotate_figure(egg::ggarrange(pwb1, pst1, ppa1, pfl1, pye1, psn1, pdb1, ncol = 1), left = "log(Yield, mm/day)")
dev.off()Plot time series just for summer 2020, for more detailed view
polycol <- "grey90"
mycols <- brewer.pal(7, "Set2")
### WEST BROOK
tempdat <- dat_clean %>%
filter(basin == "West Brook", date >= date("2020-07-01"), date <= date("2020-09-30")) %>%
mutate(site_name = factor(site_name, levels = wborder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "West Brook", date >= date("2020-07-01"), date <= date("2020-09-30"))
# same yaxis scale as main time series
pwb1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = wborder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 7, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[1], fill = NA, linewidth = 1.5)
)
### STAUNTON RIVER
tempdat <- dat_clean %>%
filter(basin == "Staunton River", date >= date("2020-07-01"), date <= date("2020-09-30")) %>%
mutate(site_name = factor(site_name, levels = stauntorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Staunton River", , date >= date("2020-07-01"), date <= date("2020-09-30"))
# color
pst1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = stauntorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 7, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[2], fill = NA, linewidth = 1.5)
)
### PAINE RUN
tempdat <- dat_clean %>%
filter(basin == "Paine Run", date >= date("2020-07-01"), date <= date("2020-09-30")) %>%
mutate(site_name = factor(site_name, levels = paineorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Paine Run", date >= date("2020-07-01"), date <= date("2020-09-30"))
# color
ppa1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = paineorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 7, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[3], fill = NA, linewidth = 1.5)
)
### FLATHEAD
tempdat <- dat_clean %>%
filter(basin == "Flathead", date >= date("2020-07-01"), date <= date("2020-09-30")) %>%
mutate(site_name = factor(site_name, levels = flatorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Flathead", date >= date("2020-07-01"), date <= date("2020-09-30"))
# color
pfl1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = flatorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 7, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[4], fill = NA, linewidth = 1.5)
)
### YELLOWSTONE
tempdat <- dat_clean %>%
filter(basin == "Shields River", date >= date("2020-07-01"), date <= date("2020-09-30")) %>%
mutate(site_name = factor(site_name, levels = yellorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Shields River", date >= date("2020-07-01"), date <= date("2020-09-30"))
# color
pye1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = yellorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 7, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[5], fill = NA, linewidth = 1.5)
)
### SNAKE
tempdat <- dat_clean %>%
filter(basin == "Snake River", date >= date("2020-07-01"), date <= date("2020-09-30")) %>%
mutate(site_name = factor(site_name, levels = snakeorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Snake River", date >= date("2020-07-01"), date <= date("2020-09-30"))
# color
psn1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = snakeorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 7, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[6], fill = NA, linewidth = 1.5)
)
### DONNER BLITZEN
tempdat <- dat_clean %>%
filter(basin == "Donner Blitzen", date >= date("2020-07-01"), date <= date("2020-09-30")) %>%
mutate(site_name = factor(site_name, levels = donnerorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Donner Blitzen", date >= date("2020-07-01"), date <= date("2020-09-30"))
# color
pdb1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = donnerorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 7, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[7], fill = NA, linewidth = 1.5)
)
annotate_figure(egg::ggarrange(pwb1, pst1, ppa1, pfl1, pye1, psn1, pdb1, ncol = 1))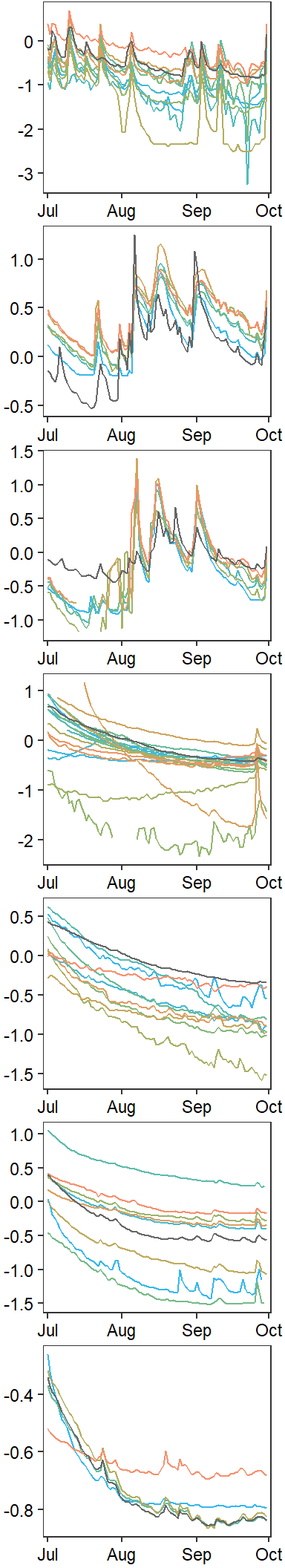
Write to file
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_timeseries_2020only.jpg", width = 2, height = 11, units = "in", res = 1000)
annotate_figure(egg::ggarrange(pwb1, pst1, ppa1, pfl1, pye1, psn1, pdb1, ncol = 1))
dev.off()Create function to plot interannual distribution of streamflow values, annual distributions of streamflow values, and scatterplot plot of
portfolioplot <- function(bas, orderr, wtryrs) {
# filter data
tempdat <- dat_clean %>%
filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% wtryrs) %>%
mutate(site_name = factor(site_name, levels = orderr))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% unique(tempdat$WaterYear))
# calculate variability for reference gage
varbig <- tempdat_big %>%
group_by(WaterYear) %>%
summarize(bigrange = range(logYield)[2]-range(logYield)[1],
bigsd = sd(logYield),
bigvar = var(logYield)) %>%
ungroup()
# interannual
pint <- ggplot() +
geom_density(data = tempdat, aes(x = logYield, y = ..scaled.., color = site_name, fill = site_name), size = 0.8) +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_fill_manual(values = alpha(cet_pal(nsites, name = "i1"), 0.1)) +
geom_density(data = tempdat_big, aes(x = logYield, y = ..scaled..), color = "grey40", fill = alpha("grey40", 0.2), size = 0.8) +
xlab("Summer log(Yield, mm/day)") + ylab("Scaled density") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.text = element_text(color = "black"), legend.position = "none")
return(pint)
}Plot distributions of summer log(yield) for all basins
# no borders
egg::ggarrange(
# West Brook
portfolioplot(bas = "West Brook", orderr = wborder, wtryrs = c(2020:2024)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 7, b = 2, l = 1)),
# Staunton
portfolioplot(bas = "Staunton River", orderr = stauntorder, wtryrs = c(2019:2022)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 7, b = 2, l = 1)),
# Paine
portfolioplot(bas = "Paine Run", orderr = paineorder, wtryrs = c(2019:2022)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 7, b = 2, l = 1)),
# Flathead
portfolioplot(bas = "Flathead", orderr = flatorder, wtryrs = c(2018:2024)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 7, b = 2, l = 1)),
# Yellowstone
portfolioplot(bas = "Shields River", orderr = yellorder, wtryrs = c(2016:2020,2022:2024)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 7, b = 2, l = 1)),
# Snake
portfolioplot(bas = "Snake River", orderr = snakeorder, wtryrs = c(2016:2024)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 7, b = 2, l = 1)),
# Donner Blitzen
portfolioplot(bas = "Donner Blitzen", orderr = donnerorder, wtryrs = c(2016:2024)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 7, b = 2, l = 1)),
nrow = 7)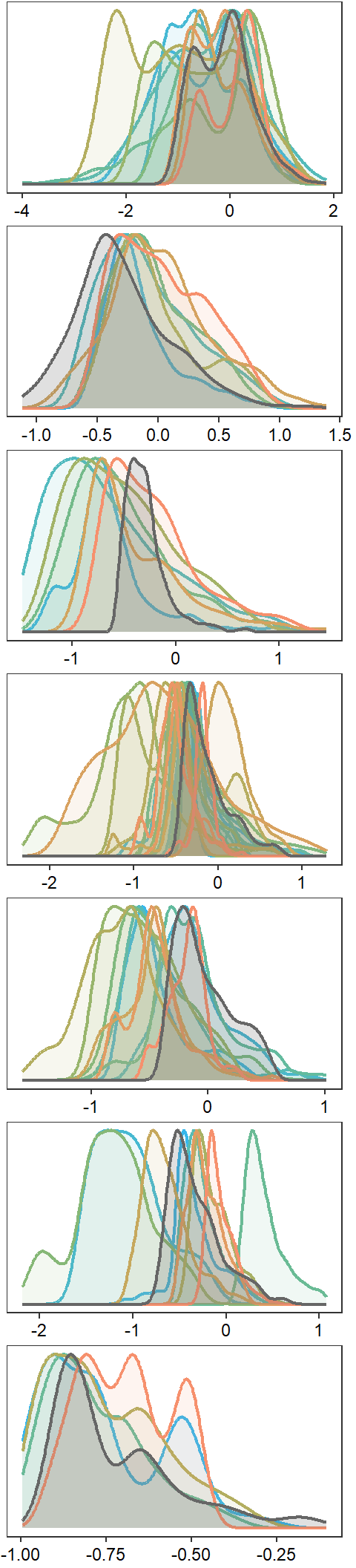
#annotate_figure(p, left = "Density")
# jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_summerdensity_all_noborder_wide.jpg", width = 2.5, height = 11, units = "in", res = 1000)
# annotate_figure(p1, left = "Density")
# dev.off()Time series plots, West Brook and Flathead
### WEST BROOK
tempdat <- dat_clean %>%
filter(basin == "West Brook") %>%
mutate(site_name = factor(site_name, levels = wborder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "West Brook", date >= date("2020-01-01"), date <= date("2025-01-03"))
# color
pwb1 <- ggplot() +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2023-07-01"), xmax = ymd("2023-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2024-07-01"), xmax = ymd("2024-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = wborder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
ylim(-3,2) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "West Brook, MA", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[1], fill = NA, linewidth = 1.5)
)
### FLATHEAD
tempdat <- dat_clean %>%
filter(basin == "Flathead", date >= date("2018-07-29"), date <= date("2023-08-11")) %>%
mutate(site_name = factor(site_name, levels = flatorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Flathead", date >= date("2018-07-29"), date <= date("2023-08-11"))
# color
pfl1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = flatorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
ylim(-3,2) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Flathead River", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
p <- annotate_figure(egg::ggarrange(pwb1, pfl1 + ylim(-2.4,1.72), ncol = 1), left = "log(Yield, mm/day)")
print(p)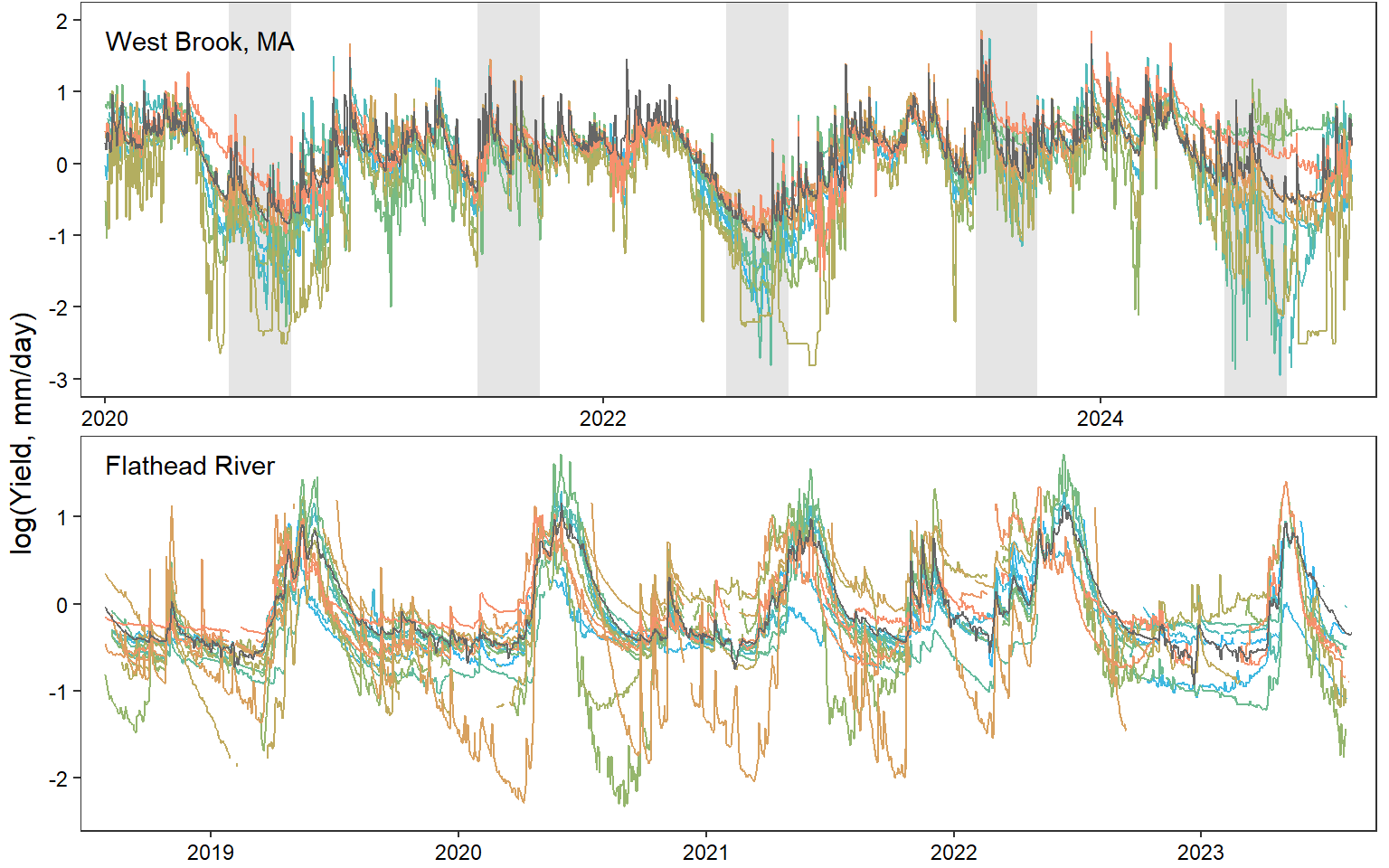
# jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_timeseries_WBFlat.jpg", width = 8, height = 5, units = "in", res = 1000)
# p
# dev.off()No headwater gages
### WEST BROOK
tempdat <- dat_clean %>%
filter(basin == "West Brook") %>%
mutate(site_name = factor(site_name, levels = wborder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "West Brook", date >= date("2020-01-01"), date <= date("2025-01-03"))
# color
pwb1_nlg <- ggplot() +
#geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = wborder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
ylim(-3,2) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "The West Brook", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### FLATHEAD
tempdat <- dat_clean %>%
filter(basin == "Flathead", date >= date("2018-07-29"), date <= date("2023-08-11")) %>%
mutate(site_name = factor(site_name, levels = flatorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Flathead", date >= date("2018-07-29"), date <= date("2023-08-11"))
# color
pfl1_nlg <- ggplot() +
#geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = flatorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
ylim(-2.4,1.72) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Flathead River", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
p <- annotate_figure(egg::ggarrange(pwb1_nlg, pfl1_nlg, ncol = 1), left = "log(Yield, mm/day)")
print(p)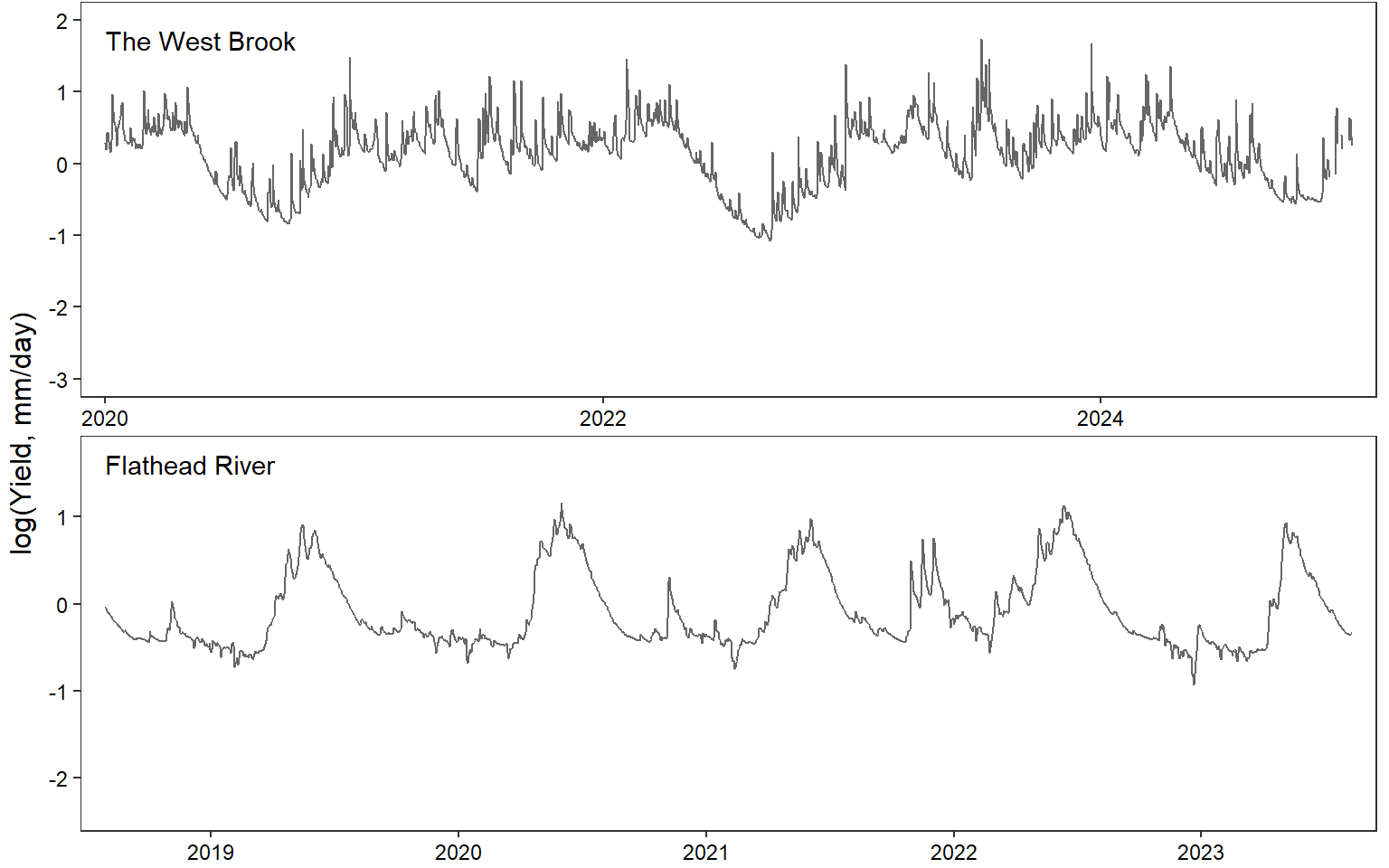
# jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_timeseries_WBFlat_nolittle.jpg", width = 8, height = 5, units = "in", res = 1000)
# p
# dev.off()Summer log(yield) distributions for West Brook and Flathead
egg::ggarrange(portfolioplot(bas = "West Brook", orderr = wborder, wtryrs = c(2020:2024)) + theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(), plot.margin = margin(t = 1, r = 1, b = 2, l = 1)),
portfolioplot(bas = "Flathead", orderr = flatorder, wtryrs = c(2018:2024)) + theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(), plot.margin = margin(t = 1, r = 1, b = 2, l = 1)),
nrow = 2)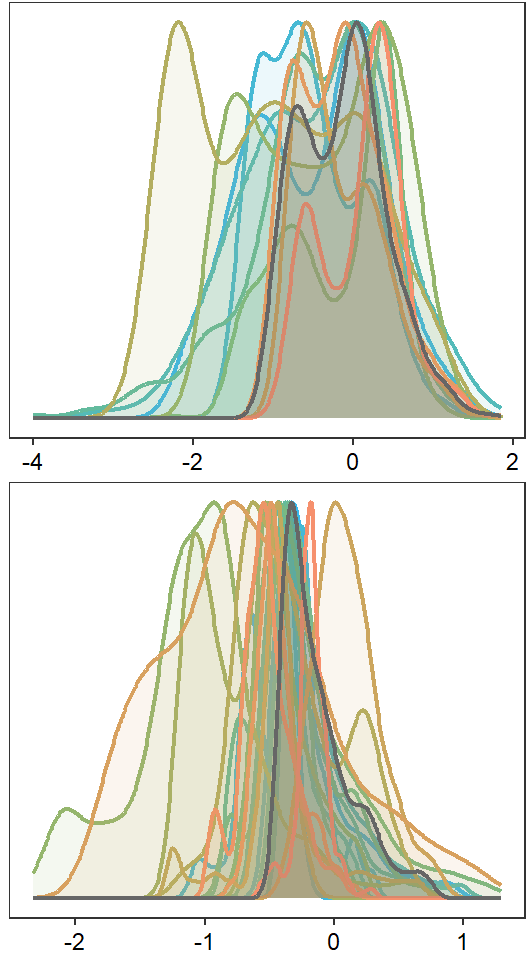
# print(p)
# jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_summerdensity_WBFlat.jpg", width = 2.75, height = 5, units = "in", res = 1000)
# p
# dev.off()How repeatable is summer monthly mean flow among sites within headwater networks? I.e., are sites with higher than average yield always higher than average, or do “lines cross” frequently?
Calculate reputability (R) using the rptR package (Stoffel et al. 2017). Also see related papers by Nakagawa and Schielzeth. Repeatibility is calculated on mean monthly flow z-scored by month and year, which removes the effect of monthly and interannual variation in water availability. Results are very similar to models fit using raw yield data with big G flow as a fixed effect.
basins <- c("West Brook", "Paine Run", "Staunton River", "Flathead", "Shields River", "Snake River", "Donner Blitzen")
rptlist_summer <- list()
rptlist_summer_cov <- list()
# summer only
for (i in 1:length(basins)) {
# daily data
td <- dat_clean %>% filter(basin == basins[i], Month %in% c(7:9))
# summarize by month
td2 <- td %>%
group_by(subbasin, site_name, designation, CalendarYear, Month, MonthName) %>%
summarize(logYield = mean(logYield)) %>%
ungroup()
# big g data
tempdat_big <- dat_clean_big %>%
filter(basin == basins[i], date >= min(td$date), date <= max(td$date)) %>%
group_by(site_name, CalendarYear, Month, MonthName) %>%
summarize(logYield = mean(logYield)) %>%
ungroup()
# join data
joined <- td2 %>%
left_join(tempdat_big %>% select(CalendarYear, MonthName, logYield) %>% rename(logYield_big = logYield)) %>%
group_by(CalendarYear, Month) %>%
mutate(logYield_z = scale(logYield)) %>%
ungroup()
# calculate repeatability
rpt_znull <- rptGaussian(logYield_z ~ 1 + (1|site_name), data = joined, grname = "site_name")
rpt_cov <- rptGaussian(logYield ~ logYield_big + (1|site_name), data = joined, grname = "site_name")
# store in list
rptlist_summer[[i]] <- tibble(basin = basins[i], rboot = rpt_znull$R_boot$site_name, type = "zscore")
rptlist_summer_cov[[i]] <- tibble(basin = basins[i], rboot = rpt_cov$R_boot$site_name, type = "cov")
}
rpttib_summer <- bind_rows(do.call(rbind, rptlist_summer), do.call(rbind, rptlist_summer_cov))Generally, repeatibility appears to be greater for snowmelt-dominated basins, particularly in the summer. The “all data” plot is more difficult to interpret as we mostly have seasonal data in the snowmelt basins, particularly the Yellowstone and Snake. In actuality, I would expect the opposite pattern if we had complete time series data for all basins: low reputability for snowmelt basins and high repeatability for rainfall basins (for the same reasons as bow tie vs. wedge).
rpttib_summer %>%
filter(basin != "Donner Blitzen", type == "zscore") %>%
mutate(basin = ifelse(basin == "Paine Run", "Paine",
ifelse(basin == "Staunton River", "Staunton",
ifelse(basin == "Snake River", "Snake",
ifelse(basin == "Shields River", "Yellowstone", basin))))) %>%
mutate(basin = factor(basin, levels = rev(c("West Brook", "Staunton", "Paine", "Flathead", "Yellowstone", "Snake")))) %>%
ggplot(aes(x = rboot, y = basin)) +
geom_density_ridges(quantile_lines = TRUE, quantiles = 2, aes(fill = basin)) +
scale_fill_manual(values = rev(brewer.pal(6, "Set2"))) +
xlim(0,1) +
theme_bw() + theme(legend.position = "none", panel.grid.major = element_blank(), panel.grid.minor = element_blank(), axis.text = element_text(color = "black")) +
xlab("Bootstrapped repeatability (R)") + ylab("Basin") + #ggtitle("Summer only (JAS)") +
geom_text(data = rpttib_summer %>% filter(basin != "Donner Blitzen", type == "zscore") %>%
mutate(basin = ifelse(basin == "Paine Run", "Paine",
ifelse(basin == "Staunton River", "Staunton",
ifelse(basin == "Snake River", "Snake",
ifelse(basin == "Shields River", "Yellowstone", basin)))))%>% group_by(basin) %>% summarise(rboot = median(rboot)),
aes(label = round(rboot, digits = 2)), position = position_nudge(y = -0.15), colour = "black", size=3.5) 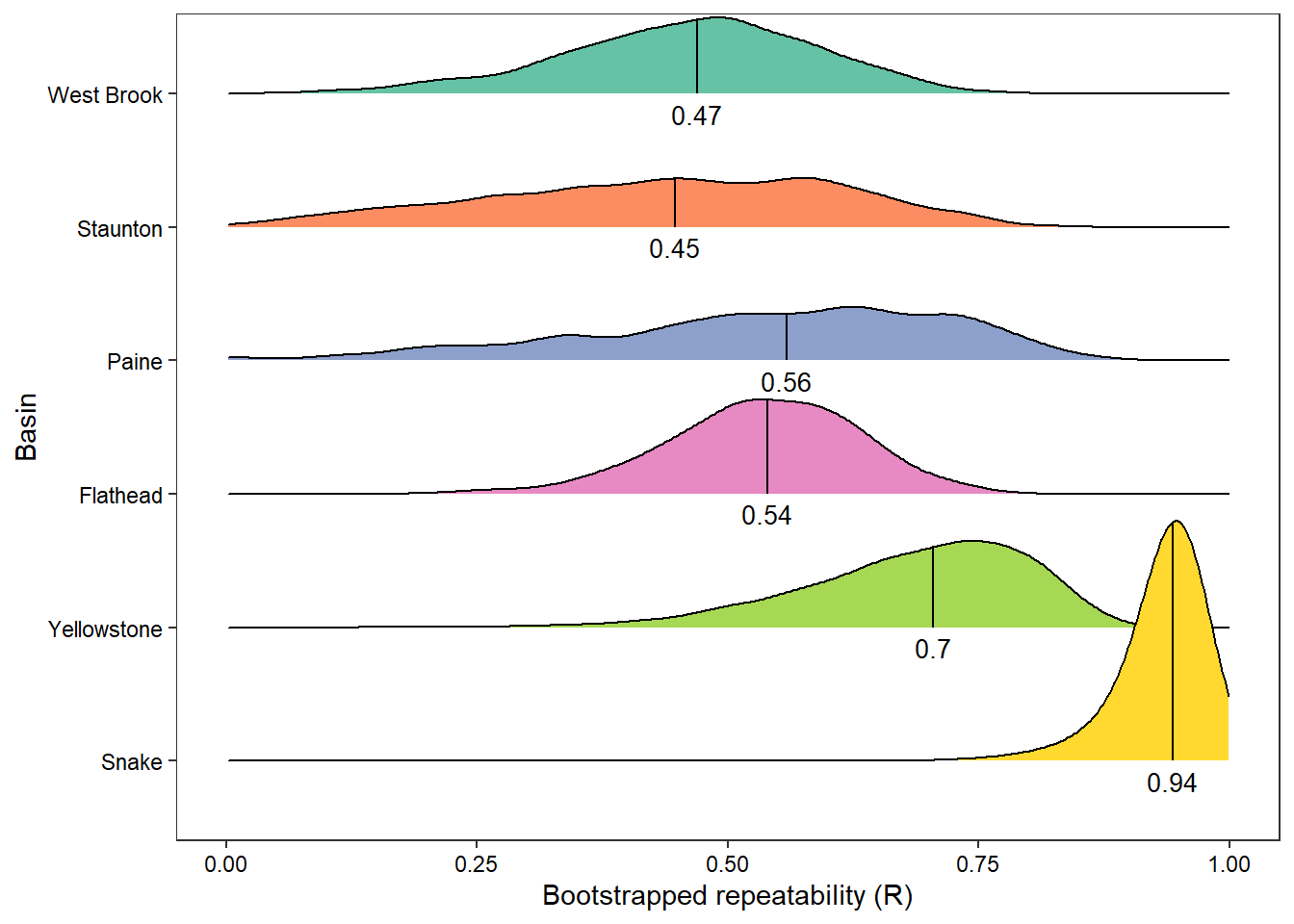
Write summer repeatability figure to file
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/MeanMonthlyFlow_SummerRepeatability.jpg", width = 4, height = 3.5, units = "in", res = 1000)
rpttib_summer %>%
filter(basin != "Donner Blitzen", type == "zscore") %>%
mutate(basin = ifelse(basin == "Paine Run", "Paine",
ifelse(basin == "Staunton River", "Staunton",
ifelse(basin == "Snake River", "Snake",
ifelse(basin == "Shields River", "Yellowstone", basin))))) %>%
mutate(basin = factor(basin, levels = rev(c("West Brook", "Staunton", "Paine", "Flathead", "Yellowstone", "Snake")))) %>%
ggplot(aes(x = rboot, y = basin)) +
geom_density_ridges(quantile_lines = TRUE, quantiles = 2, aes(fill = basin)) +
scale_fill_manual(values = rev(brewer.pal(6, "Set2"))) +
xlim(0,1) +
theme_bw() + theme(legend.position = "none", panel.grid.major = element_blank(), panel.grid.minor = element_blank(), axis.text = element_text(color = "black")) +
xlab("Bootstrapped repeatability (R)") + ylab("Basin") + #ggtitle("Summer only (JAS)") +
geom_text(data = rpttib_summer %>% filter(basin != "Donner Blitzen", type == "zscore") %>%
mutate(basin = ifelse(basin == "Paine Run", "Paine",
ifelse(basin == "Staunton River", "Staunton",
ifelse(basin == "Snake River", "Snake",
ifelse(basin == "Shields River", "Yellowstone", basin)))))%>% group_by(basin) %>% summarise(rboot = median(rboot)),
aes(label = round(rboot, digits = 2)), position = position_nudge(y = -0.15), colour = "black", size=3.5)
dev.off()Use modified fluvial synchrograms to graphically explore the geography of spatial heterogeneity in summer streamflow (sensu Larsen et al. 2018 Ecology Letters). How does time series similarity (distance) among headwater sites change as a function of euclidean, watercourse, and flow-connected geographic distance? Given spatial dependencies in river networks (e.g., Peterson et al. 2013 Ecology Letters), we would expect flow regimes to be more similar among flow-connected sites located in close proximity (i.e., increasing time series distance with increasing geographic distance).
Crop flow line to area of interest:
# West Brook
mysheds <- read_sf(dsn = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds", layer = "Mass_Watersheds")
mysheds <- mysheds[mysheds$site_id == "WBR",]
mynet <- read_sf("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams", layer = "Mass_Streams")
st_crs(mynet) <- st_crs(mysheds)
mynet <- st_intersection(mynet, mysheds)
mynet <- st_transform(mynet, crs = "+proj=utm +zone=18")
st_write(mynet, "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/FluvialSynch/Spatial/Mass_Streams_crop.shp", append = FALSE)
# Staunton
mysheds <- read_sf(dsn = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds", layer = "Shen_Watersheds")
mysheds <- mysheds[mysheds$site_id == "SR_10FL",]
mynet <- read_sf("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams", layer = "Shen_Streams")
st_crs(mynet) <- st_crs(mysheds)
mynet <- st_intersection(mynet, mysheds)
mynet <- st_transform(mynet, crs = "+proj=utm +zone=18")
st_write(mynet, "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/FluvialSynch/Spatial/Staunton_Streams_crop.shp", append = FALSE)
# Paine Run
mysheds <- read_sf(dsn = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds", layer = "Shen_Watersheds")
mysheds <- mysheds[mysheds$site_id == "PA_10FL",]
mynet <- read_sf("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams", layer = "Shen_Streams")
st_crs(mynet) <- st_crs(mysheds)
mynet <- st_intersection(mynet, mysheds)
mynet <- st_transform(mynet, crs = "+proj=utm +zone=18")
st_write(mynet, "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/FluvialSynch/Spatial/Paine_Streams_crop.shp", append = FALSE)
# Flathead
mysheds <- read_sf(dsn = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds", layer = "Flat_Watersheds")
mysheds <- mysheds[mysheds$site_id == "NFF",]
mynet <- read_sf("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams", layer = "Flat_Streams")
st_crs(mynet) <- st_crs(mysheds)
mynet <- st_intersection(mynet, mysheds)
mynet <- st_transform(mynet, crs = "+proj=utm +zone=18")
st_write(mynet, "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/FluvialSynch/Spatial/Flat_Streams_crop.shp", append = FALSE)
# Yellowstone
mysheds <- read_sf(dsn = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds", layer = "Shields_Watersheds")
mysheds <- mysheds[mysheds$site_id == "SRS",]
mynet <- read_sf("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams", layer = "Shields_Streams")
st_crs(mynet) <- st_crs(mysheds)
mynet <- st_intersection(mynet, mysheds)
mynet <- st_transform(mynet, crs = "+proj=utm +zone=18")
st_write(mynet, "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/FluvialSynch/Spatial/Yell_Streams_crop.shp", append = FALSE)
# Snake
mysheds <- read_sf(dsn = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds", layer = "Snake_Watersheds")
mysheds <- mysheds[mysheds$site_id == "SP11",]
mynet <- read_sf("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams", layer = "Snake_Streams")
st_crs(mynet) <- st_crs(mysheds)
mynet <- st_intersection(mynet, mysheds)
mynet <- st_transform(mynet, crs = "+proj=utm +zone=18")
st_write(mynet, "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/FluvialSynch/Spatial/Snake_Streams_crop.shp", append = FALSE)
# Donner-Blitzen
mysheds <- read_sf(dsn = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds", layer = "Oreg_Watersheds")
mysheds <- mysheds[mysheds$site_id == "DBF",]
mynet <- read_sf("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams", layer = "Oreg_Streams")
st_crs(mynet) <- st_crs(mysheds)
mynet <- st_intersection(mynet, mysheds)
mynet <- st_transform(mynet, crs = "+proj=utm +zone=18")
st_write(mynet, "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/FluvialSynch/Spatial/DonnerBlitzen_Streams_crop.shp", append = FALSE)Convert flowline to riverdist network object:
# West Brook
flowline <- line2network(path = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/FluvialSynch/Spatial", layer = "Mass_Streams_crop")
Units: metre
Removed 2 segments with lengths shorter than the connectivity tolerance. # plot(flowline)
# zoomtoseg(seg = c(4), rivers = flowline)
# showends(seg = 4, rivers = flowline)
flowline_wb <- setmouth(seg = 4, vert = 1, rivers = flowline)
# Staunton
flowline <- line2network(path = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/FluvialSynch/Spatial", layer = "Staunton_Streams_crop")
Units: metre # plot(flowline)
# zoomtoseg(seg = c(2), rivers = flowline)
# showends(seg = 2, rivers = flowline)
flowline_st <- setmouth(seg = 2, vert = 29, rivers = flowline)
# Paine
flowline <- line2network(path = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/FluvialSynch/Spatial", layer = "Paine_Streams_crop")
Units: metre
Removed 1 segments with lengths shorter than the connectivity tolerance. # plot(flowline)
# zoomtoseg(seg = c(6), rivers = flowline)
# showends(seg = 6, rivers = flowline)
flowline_pa <- setmouth(seg = 6, vert = 23, rivers = flowline)
# Flathead
flowline <- line2network(path = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/FluvialSynch/Spatial", layer = "Flat_Streams_crop")
Units: metre
Removed 28 segments with lengths shorter than the connectivity tolerance. # plot(flowline)
# zoomtoseg(seg = c(6, 46, 59, 7), rivers = flowline)
# showends(seg = 7, rivers = flowline)
flowline_fl <- setmouth(seg = 7, vert = 82, rivers = flowline)
# Yellowstone
flowline <- line2network(path = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/FluvialSynch/Spatial", layer = "Yell_Streams_crop")
Units: metre
Removed 2 segments with lengths shorter than the connectivity tolerance. # plot(flowline)
# zoomtoseg(seg = c(6), rivers = flowline)
# showends(seg = 6, rivers = flowline)
flowline_ye <- setmouth(seg = 6, vert = 197, rivers = flowline)
# Snake
flowline <- line2network(path = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/FluvialSynch/Spatial", layer = "Snake_Streams_crop")
Units: metre
Removed 3 segments with lengths shorter than the connectivity tolerance. # plot(flowline)
# zoomtoseg(seg = c(22), rivers = flowline)
# showends(seg = 22, rivers = flowline)
flowline_sn <- setmouth(seg = 22, vert = 88, rivers = flowline)
# Donner Blitzen
flowline <- line2network(path = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/FluvialSynch/Spatial", layer = "DonnerBlitzen_Streams_crop")
Units: metre
Removed 4 segments with lengths shorter than the connectivity tolerance. # plot(flowline)
# zoomtoseg(seg = c(37,206, 209, 38), rivers = flowline)
# showends(seg = 38, rivers = flowline)
flowline_db <- setmouth(seg = 38, vert = 20, rivers = flowline)Convert focal sites to riverpoints, snapped to networks
mysites_wb <- siteinfo %>% filter(subbasin == "West Brook", site_name %in% unique(dat_clean$site_name))
mysites_wb_sp <- st_as_sf(mysites_wb, coords = c("long", "lat"), crs = 4326)
mysites_wb_sp <- st_transform(mysites_wb_sp, crs = st_crs(mynet))
mysites_wb_sp_xy <- xy2segvert(x = st_coordinates(mysites_wb_sp)[,1], y = st_coordinates(mysites_wb_sp)[,2], rivers = flowline_wb)
plot(flowline_wb)
riverpoints(seg = mysites_wb_sp_xy$seg, vert = mysites_wb_sp_xy$vert, rivers = flowline_wb, pch = 16, col = "blue")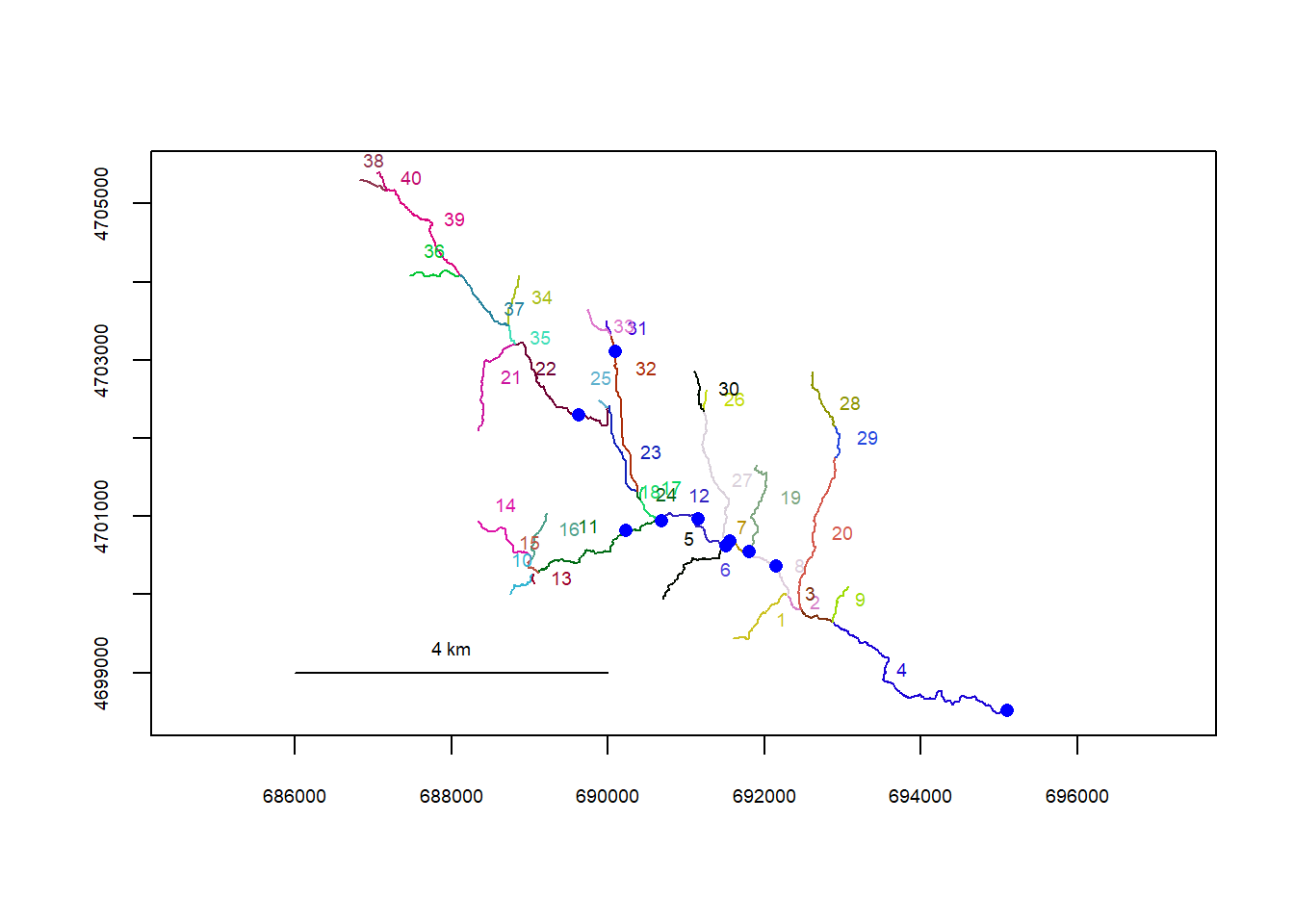
mysites_st <- siteinfo %>% filter(subbasin == "Staunton River", site_name %in% unique(dat_clean$site_name))
mysites_st_sp <- st_as_sf(mysites_st, coords = c("long", "lat"), crs = 4326)
mysites_st_sp <- st_transform(mysites_st_sp, crs = st_crs(mynet))
mysites_st_sp_xy <- xy2segvert(x = st_coordinates(mysites_st_sp)[,1], y = st_coordinates(mysites_st_sp)[,2], rivers = flowline_st)
plot(flowline_st)
riverpoints(seg = mysites_st_sp_xy$seg, vert = mysites_st_sp_xy$vert, rivers = flowline_st, pch = 16, col = "blue")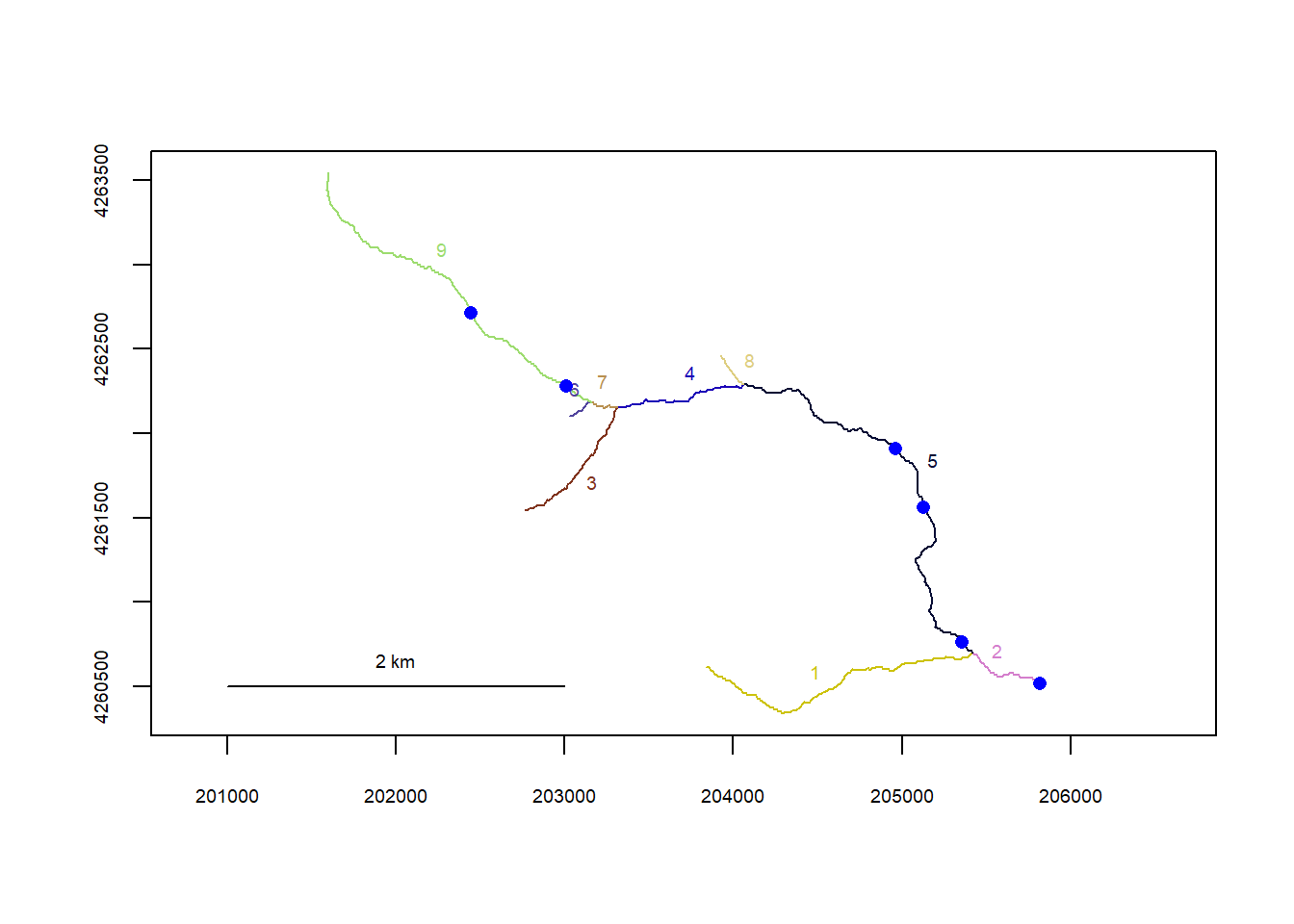
mysites_pa <- siteinfo %>% filter(subbasin == "Paine Run", site_name %in% unique(dat_clean$site_name))
mysites_pa_sp <- st_as_sf(mysites_pa, coords = c("long", "lat"), crs = 4326)
mysites_pa_sp <- st_transform(mysites_pa_sp, crs = st_crs(mynet))
mysites_pa_sp_xy <- xy2segvert(x = st_coordinates(mysites_pa_sp)[,1], y = st_coordinates(mysites_pa_sp)[,2], rivers = flowline_pa)
plot(flowline_pa)
riverpoints(seg = mysites_pa_sp_xy$seg, vert = mysites_pa_sp_xy$vert, rivers = flowline_pa, pch = 16, col = "blue")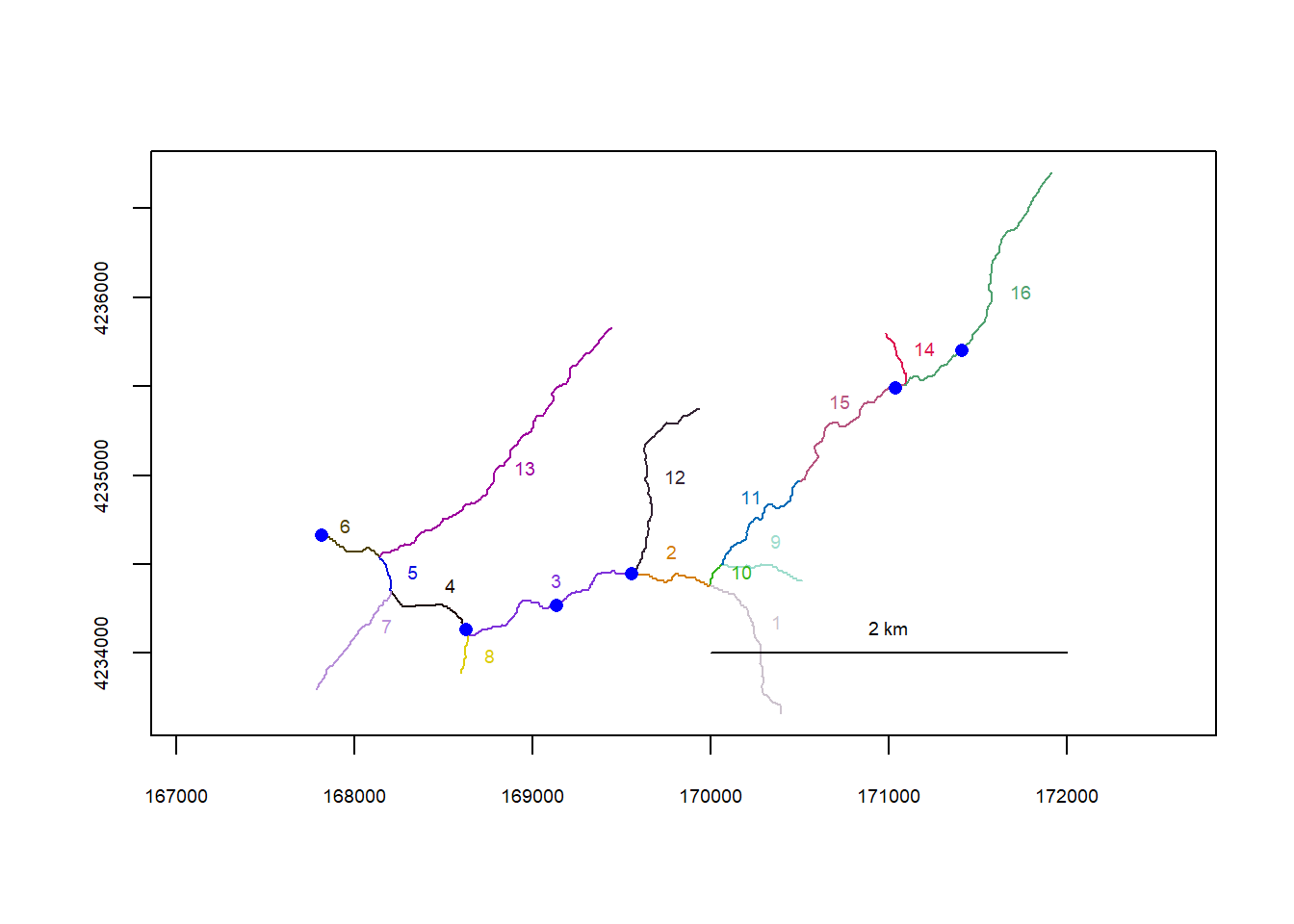
mysites_fl <- siteinfo %>% filter(basin == "Flathead", site_name %in% unique(dat_clean$site_name))
mysites_fl_sp <- st_as_sf(mysites_fl, coords = c("long", "lat"), crs = 4326)
mysites_fl_sp <- st_transform(mysites_fl_sp, crs = st_crs(mynet))
mysites_fl_sp_xy <- xy2segvert(x = st_coordinates(mysites_fl_sp)[,1], y = st_coordinates(mysites_fl_sp)[,2], rivers = flowline_fl)
plot(flowline_fl)
riverpoints(seg = mysites_fl_sp_xy$seg, vert = mysites_fl_sp_xy$vert, rivers = flowline_fl, pch = 16, col = "blue")
mysites_ye <- siteinfo %>% filter(subbasin == "Shields River", site_name %in% unique(dat_clean$site_name))
mysites_ye_sp <- st_as_sf(mysites_ye, coords = c("long", "lat"), crs = 4326)
mysites_ye_sp <- st_transform(mysites_ye_sp, crs = st_crs(mynet))
mysites_ye_sp_xy <- xy2segvert(x = st_coordinates(mysites_ye_sp)[,1], y = st_coordinates(mysites_ye_sp)[,2], rivers = flowline_ye)
plot(flowline_ye)
riverpoints(seg = mysites_ye_sp_xy$seg, vert = mysites_ye_sp_xy$vert, rivers = flowline_ye, pch = 16, col = "blue")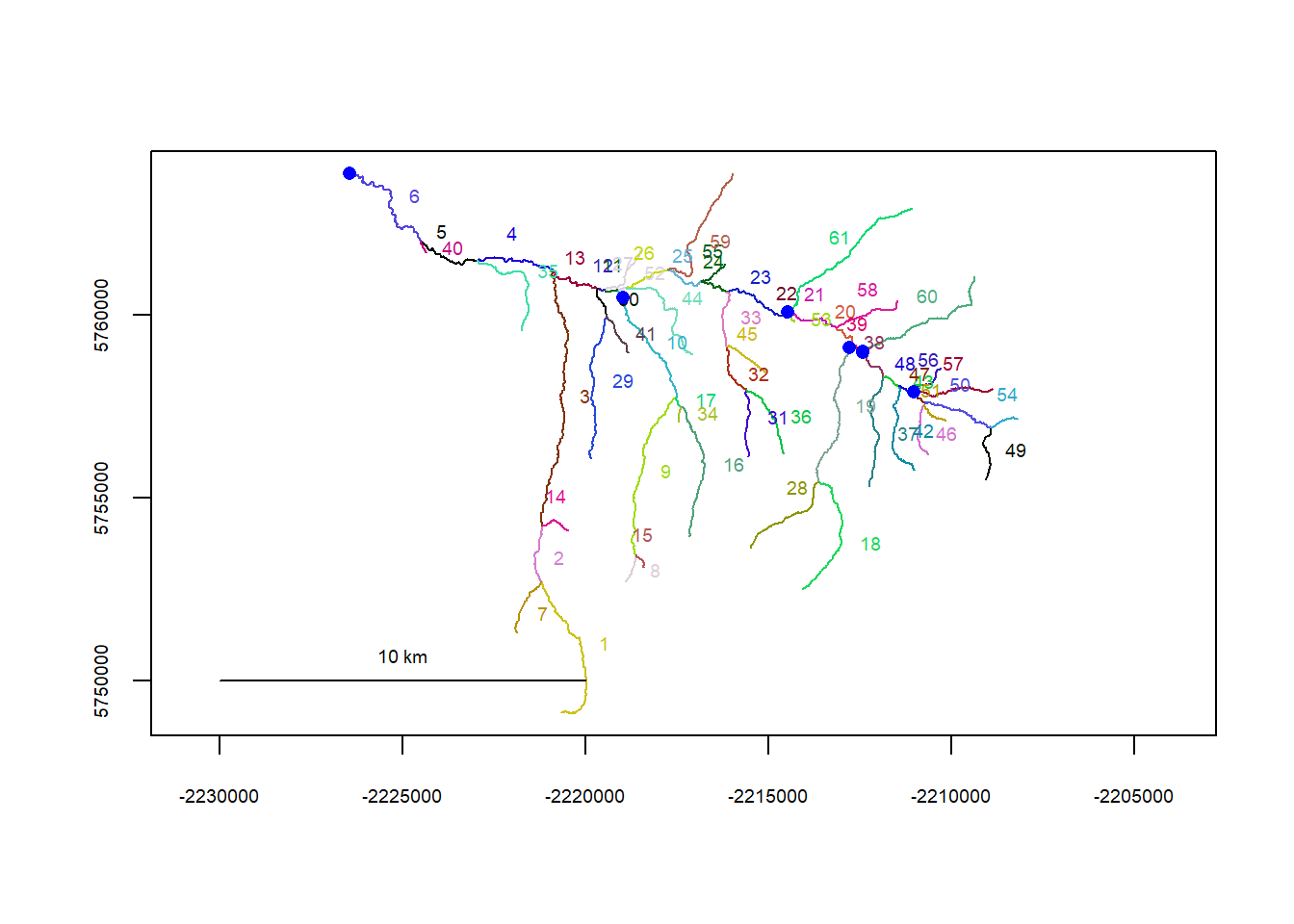
mysites_sn <- siteinfo %>% filter(subbasin == "Snake River", site_name %in% unique(dat_clean$site_name))
mysites_sn_sp <- st_as_sf(mysites_sn, coords = c("long", "lat"), crs = 4326)
mysites_sn_sp <- st_transform(mysites_sn_sp, crs = st_crs(mynet))
mysites_sn_sp_xy <- xy2segvert(x = st_coordinates(mysites_sn_sp)[,1], y = st_coordinates(mysites_sn_sp)[,2], rivers = flowline_sn)
plot(flowline_sn)
riverpoints(seg = mysites_sn_sp_xy$seg, vert = mysites_sn_sp_xy$vert, rivers = flowline_sn, pch = 16, col = "blue")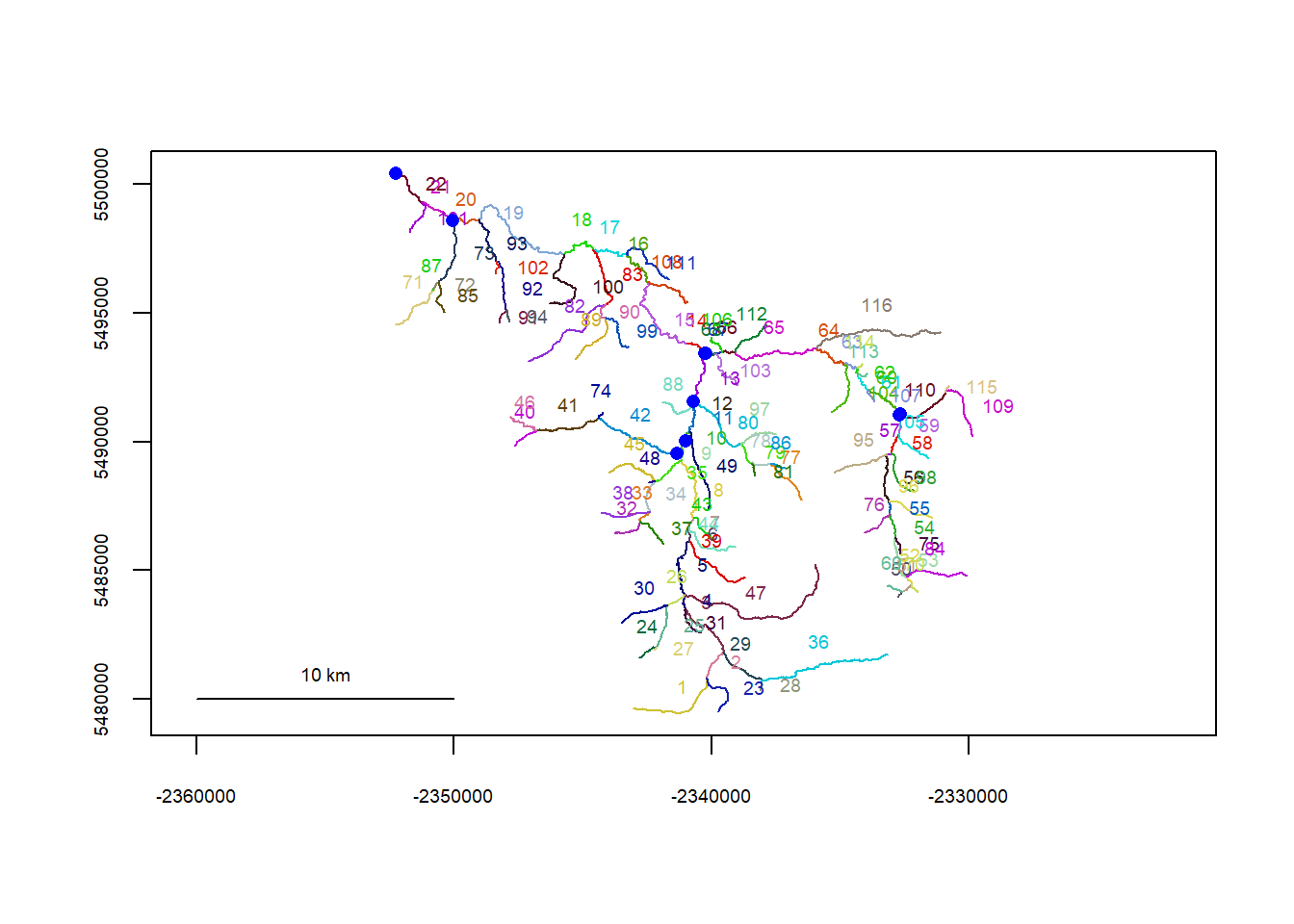
mysites_db <- siteinfo %>% filter(subbasin == "Donner Blitzen", site_name %in% unique(dat_clean$site_name))
mysites_db_sp <- st_as_sf(mysites_db, coords = c("long", "lat"), crs = 4326)
mysites_db_sp <- st_transform(mysites_db_sp, crs = st_crs(mynet))
mysites_db_sp_xy <- xy2segvert(x = st_coordinates(mysites_db_sp)[,1], y = st_coordinates(mysites_db_sp)[,2], rivers = flowline_db)
plot(flowline_db)
riverpoints(seg = mysites_db_sp_xy$seg, vert = mysites_db_sp_xy$vert, rivers = flowline_db, pch = 16, col = "blue")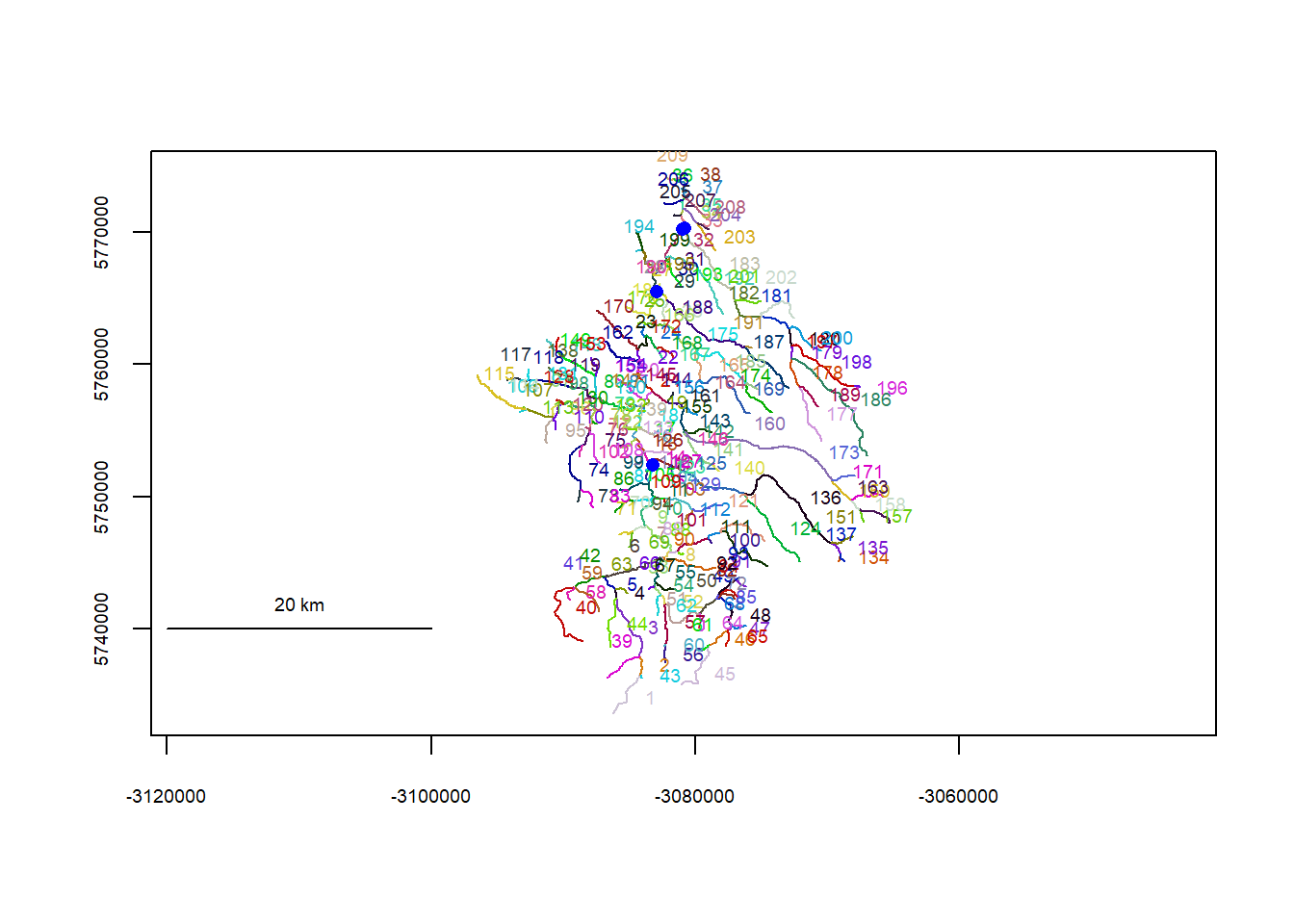
Calculate pairwise geographic distances.
# West Brook
dist_euclidean_wb <- (st_distance(mysites_wb_sp, mysites_wb_sp)) / 1000
attr(dist_euclidean_wb, "units") <- NULL
row.names(dist_euclidean_wb) <- mysites_wb$site_name
colnames(dist_euclidean_wb) <- mysites_wb$site_name
dist_euclidean_wb[upper.tri(dist_euclidean_wb, diag = TRUE)] <- NA
class(dist_euclidean_wb) <- "matrix"
# dist_euclidean_wb
# Stuanton
dist_euclidean_st <- (st_distance(mysites_st_sp, mysites_st_sp)) / 1000
attr(dist_euclidean_st, "units") <- NULL
row.names(dist_euclidean_st) <- mysites_st$site_name
colnames(dist_euclidean_st) <- mysites_st$site_name
dist_euclidean_st[upper.tri(dist_euclidean_st, diag = TRUE)] <- NA
class(dist_euclidean_st) <- "matrix"
# dist_euclidean_st
# Paine
dist_euclidean_pa <- (st_distance(mysites_pa_sp, mysites_pa_sp)) / 1000
attr(dist_euclidean_pa, "units") <- NULL
row.names(dist_euclidean_pa) <- mysites_pa$site_name
colnames(dist_euclidean_pa) <- mysites_pa$site_name
dist_euclidean_pa[upper.tri(dist_euclidean_pa, diag = TRUE)] <- NA
class(dist_euclidean_pa) <- "matrix"
# dist_euclidean_pa
# Flathead
dist_euclidean_fl <- (st_distance(mysites_fl_sp, mysites_fl_sp)) / 1000
attr(dist_euclidean_fl, "units") <- NULL
row.names(dist_euclidean_fl) <- mysites_fl$site_name
colnames(dist_euclidean_fl) <- mysites_fl$site_name
dist_euclidean_fl[upper.tri(dist_euclidean_fl, diag = TRUE)] <- NA
class(dist_euclidean_fl) <- "matrix"
# dist_euclidean_fl
# Yellowstone
dist_euclidean_ye <- (st_distance(mysites_ye_sp, mysites_ye_sp)) / 1000
attr(dist_euclidean_ye, "units") <- NULL
row.names(dist_euclidean_ye) <- mysites_ye$site_name
colnames(dist_euclidean_ye) <- mysites_ye$site_name
dist_euclidean_ye[upper.tri(dist_euclidean_ye, diag = TRUE)] <- NA
class(dist_euclidean_ye) <- "matrix"
# dist_euclidean_ye
# Snake
dist_euclidean_sn <- (st_distance(mysites_sn_sp, mysites_sn_sp)) / 1000
attr(dist_euclidean_sn, "units") <- NULL
row.names(dist_euclidean_sn) <- mysites_sn$site_name
colnames(dist_euclidean_sn) <- mysites_sn$site_name
dist_euclidean_sn[upper.tri(dist_euclidean_sn, diag = TRUE)] <- NA
class(dist_euclidean_sn) <- "matrix"
# dist_euclidean_sn
# Donner Blitzen
dist_euclidean_db <- (st_distance(mysites_db_sp, mysites_db_sp)) / 1000
attr(dist_euclidean_db, "units") <- NULL
row.names(dist_euclidean_db) <- mysites_db$site_name
colnames(dist_euclidean_db) <- mysites_db$site_name
dist_euclidean_db[upper.tri(dist_euclidean_db, diag = TRUE)] <- NA
class(dist_euclidean_db) <- "matrix"
# dist_euclidean_db# West Brook
dist_watercourse_wb <- riverdistancemat(mysites_wb_sp_xy$seg, mysites_wb_sp_xy$vert, flowline_wb) / 1000
row.names(dist_watercourse_wb) <- mysites_wb$site_name
colnames(dist_watercourse_wb) <- mysites_wb$site_name
dist_watercourse_wb[upper.tri(dist_watercourse_wb, diag = TRUE)] <- NA
# dist_watercourse_wb
# Staunton
dist_watercourse_st <- riverdistancemat(mysites_st_sp_xy$seg, mysites_st_sp_xy$vert, flowline_st) / 1000
row.names(dist_watercourse_st) <- mysites_st$site_name
colnames(dist_watercourse_st) <- mysites_st$site_name
dist_watercourse_st[upper.tri(dist_watercourse_st, diag = TRUE)] <- NA
# dist_watercourse_st
# Paine
dist_watercourse_pa <- riverdistancemat(mysites_pa_sp_xy$seg, mysites_pa_sp_xy$vert, flowline_pa) / 1000
row.names(dist_watercourse_pa) <- mysites_pa$site_name
colnames(dist_watercourse_pa) <- mysites_pa$site_name
dist_watercourse_pa[upper.tri(dist_watercourse_pa, diag = TRUE)] <- NA
# dist_watercourse_pa
# Flathead
dist_watercourse_fl <- riverdistancemat(mysites_fl_sp_xy$seg, mysites_fl_sp_xy$vert, flowline_fl) / 1000
row.names(dist_watercourse_fl) <- mysites_fl$site_name
colnames(dist_watercourse_fl) <- mysites_fl$site_name
dist_watercourse_fl[upper.tri(dist_watercourse_fl, diag = TRUE)] <- NA
# dist_watercourse_fl
# Yellowstone
dist_watercourse_ye <- riverdistancemat(mysites_ye_sp_xy$seg, mysites_ye_sp_xy$vert, flowline_ye) / 1000
row.names(dist_watercourse_ye) <- mysites_ye$site_name
colnames(dist_watercourse_ye) <- mysites_ye$site_name
dist_watercourse_ye[upper.tri(dist_watercourse_ye, diag = TRUE)] <- NA
# dist_watercourse_ye
# Snake
dist_watercourse_sn <- riverdistancemat(mysites_sn_sp_xy$seg, mysites_sn_sp_xy$vert, flowline_sn) / 1000
row.names(dist_watercourse_sn) <- mysites_sn$site_name
colnames(dist_watercourse_sn) <- mysites_sn$site_name
dist_watercourse_sn[upper.tri(dist_watercourse_sn, diag = TRUE)] <- NA
# dist_watercourse_snake
# Donner-Blitzen
dist_watercourse_db <- riverdistancemat(mysites_db_sp_xy$seg, mysites_db_sp_xy$vert, flowline_db) / 1000
row.names(dist_watercourse_db) <- mysites_db$site_name
colnames(dist_watercourse_db) <- mysites_db$site_name
dist_watercourse_db[upper.tri(dist_watercourse_db, diag = TRUE)] <- NA
# dist_watercourse_db# West Brook
dist_flowconn_wb <- abs(upstreammat(mysites_wb_sp_xy$seg, mysites_wb_sp_xy$vert, flowline_wb, flowconnected = TRUE)) / 1000
row.names(dist_flowconn_wb) <- mysites_wb$site_name
colnames(dist_flowconn_wb) <- mysites_wb$site_name
dist_flowconn_wb[upper.tri(dist_flowconn_wb, diag = TRUE)] <- NA
# dist_flowconn_wb
# Staunton
dist_flowconn_st <- abs(upstreammat(mysites_st_sp_xy$seg, mysites_st_sp_xy$vert, flowline_st, flowconnected = TRUE)) / 1000
row.names(dist_flowconn_st) <- mysites_st$site_name
colnames(dist_flowconn_st) <- mysites_st$site_name
dist_flowconn_st[upper.tri(dist_flowconn_st, diag = TRUE)] <- NA
# dist_flowconn_st
# Paine
dist_flowconn_pa <- abs(upstreammat(mysites_pa_sp_xy$seg, mysites_pa_sp_xy$vert, flowline_pa, flowconnected = TRUE)) / 1000
row.names(dist_flowconn_pa) <- mysites_pa$site_name
colnames(dist_flowconn_pa) <- mysites_pa$site_name
dist_flowconn_pa[upper.tri(dist_flowconn_pa, diag = TRUE)] <- NA
# dist_flowconn_pa
# Flathead
dist_flowconn_fl <- abs(upstreammat(mysites_fl_sp_xy$seg, mysites_fl_sp_xy$vert, flowline_fl, flowconnected = TRUE)) / 1000
row.names(dist_flowconn_fl) <- mysites_fl$site_name
colnames(dist_flowconn_fl) <- mysites_fl$site_name
dist_flowconn_fl[upper.tri(dist_flowconn_fl, diag = TRUE)] <- NA
# dist_flowconn_fl
# Yellowstone
dist_flowconn_ye <- abs(upstreammat(mysites_ye_sp_xy$seg, mysites_ye_sp_xy$vert, flowline_ye, flowconnected = TRUE)) / 1000
row.names(dist_flowconn_ye) <- mysites_ye$site_name
colnames(dist_flowconn_ye) <- mysites_ye$site_name
dist_flowconn_ye[upper.tri(dist_flowconn_ye, diag = TRUE)] <- NA
# dist_flowconn_ye
# Snake
dist_flowconn_sn <- abs(upstreammat(mysites_sn_sp_xy$seg, mysites_sn_sp_xy$vert, flowline_sn, flowconnected = TRUE)) / 1000
row.names(dist_flowconn_sn) <- mysites_sn$site_name
colnames(dist_flowconn_sn) <- mysites_sn$site_name
dist_flowconn_sn[upper.tri(dist_flowconn_sn, diag = TRUE)] <- NA
# dist_flowconn_sn
# Donner Blitzen
dist_flowconn_db <- abs(upstreammat(mysites_db_sp_xy$seg, mysites_db_sp_xy$vert, flowline_db, flowconnected = TRUE)) / 1000
row.names(dist_flowconn_db) <- mysites_db$site_name
colnames(dist_flowconn_db) <- mysites_db$site_name
dist_flowconn_db[upper.tri(dist_flowconn_db, diag = TRUE)] <- NA
# dist_flowconn_dbRealized network/branching complexity, sensu Larsen et al. (2021)
Euclidean distance ~ watercourse distance
disttib <- tibble(basin = c(rep("West Brook", times = length(as.numeric(dist_euclidean_wb / dist_watercourse_wb))),
rep("Staunton River", times = length(as.numeric(dist_euclidean_st / dist_watercourse_st))),
rep("Paine Run", times = length(as.numeric(dist_euclidean_pa / dist_watercourse_pa))),
rep("Flathead River", times = length(as.numeric(dist_euclidean_fl / dist_watercourse_fl))),
rep("Yellowstone River", times = length(as.numeric(dist_euclidean_ye / dist_watercourse_ye))),
rep("Snake River", times = length(as.numeric(dist_euclidean_sn / dist_watercourse_sn))),
rep("Donner und Blitzen River", times = length(as.numeric(dist_euclidean_db / dist_watercourse_db)))),
dist_euc = c(as.numeric(dist_euclidean_wb),
as.numeric(dist_euclidean_st),
as.numeric(dist_euclidean_pa),
as.numeric(dist_euclidean_fl),
as.numeric(dist_euclidean_ye),
as.numeric(dist_euclidean_sn),
as.numeric(dist_euclidean_db)),
dist_wat = c(as.numeric(dist_watercourse_wb),
as.numeric(dist_watercourse_st),
as.numeric(dist_watercourse_pa),
as.numeric(dist_watercourse_fl),
as.numeric(dist_watercourse_ye),
as.numeric(dist_watercourse_sn),
as.numeric(dist_watercourse_db)))
disttib %>%
ggplot() +
geom_point(aes(x = dist_wat, y = dist_euc)) +
geom_abline(intercept = 0, slope = 1, linetype = 2) +
facet_wrap(~basin) +
xlab("Watercourse distance (km)") + ylab("Euclidean distance (km)") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())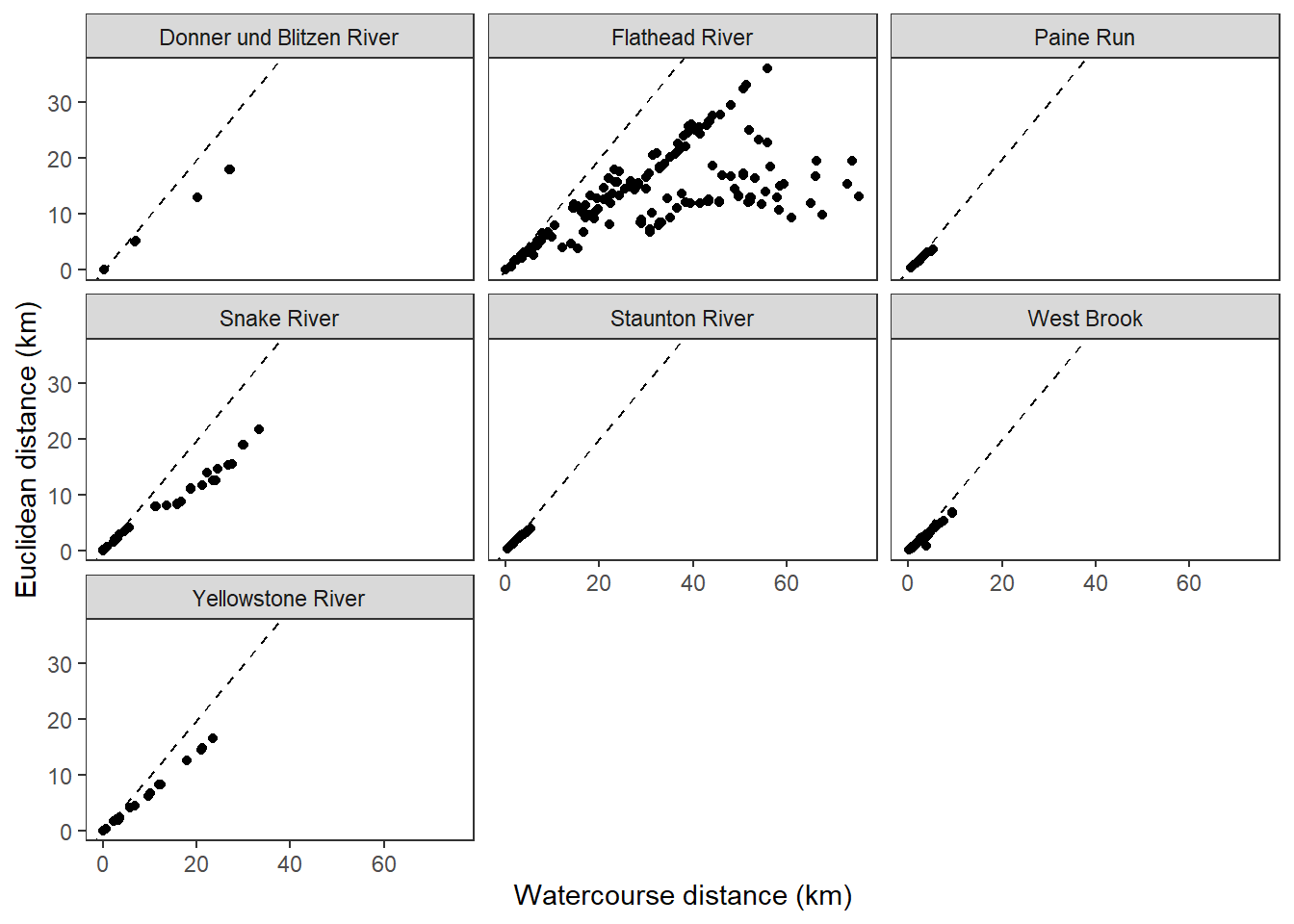
hull <- disttib %>% filter(!is.na(dist_euc), !is.na(dist_wat)) %>% group_by(basin) %>% slice(chull(dist_wat, dist_euc))
ggplot(data = disttib, aes(x = dist_wat, y = dist_euc, color = basin)) +
geom_point() +
geom_abline(intercept = 0, slope = 1, linetype = 2) +
geom_polygon(data = hull, aes(fill = basin), alpha = 0.5) +
#facet_wrap(~basin) +
xlab("Watercourse distance (km)") + ylab("Euclidean distance (km)") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())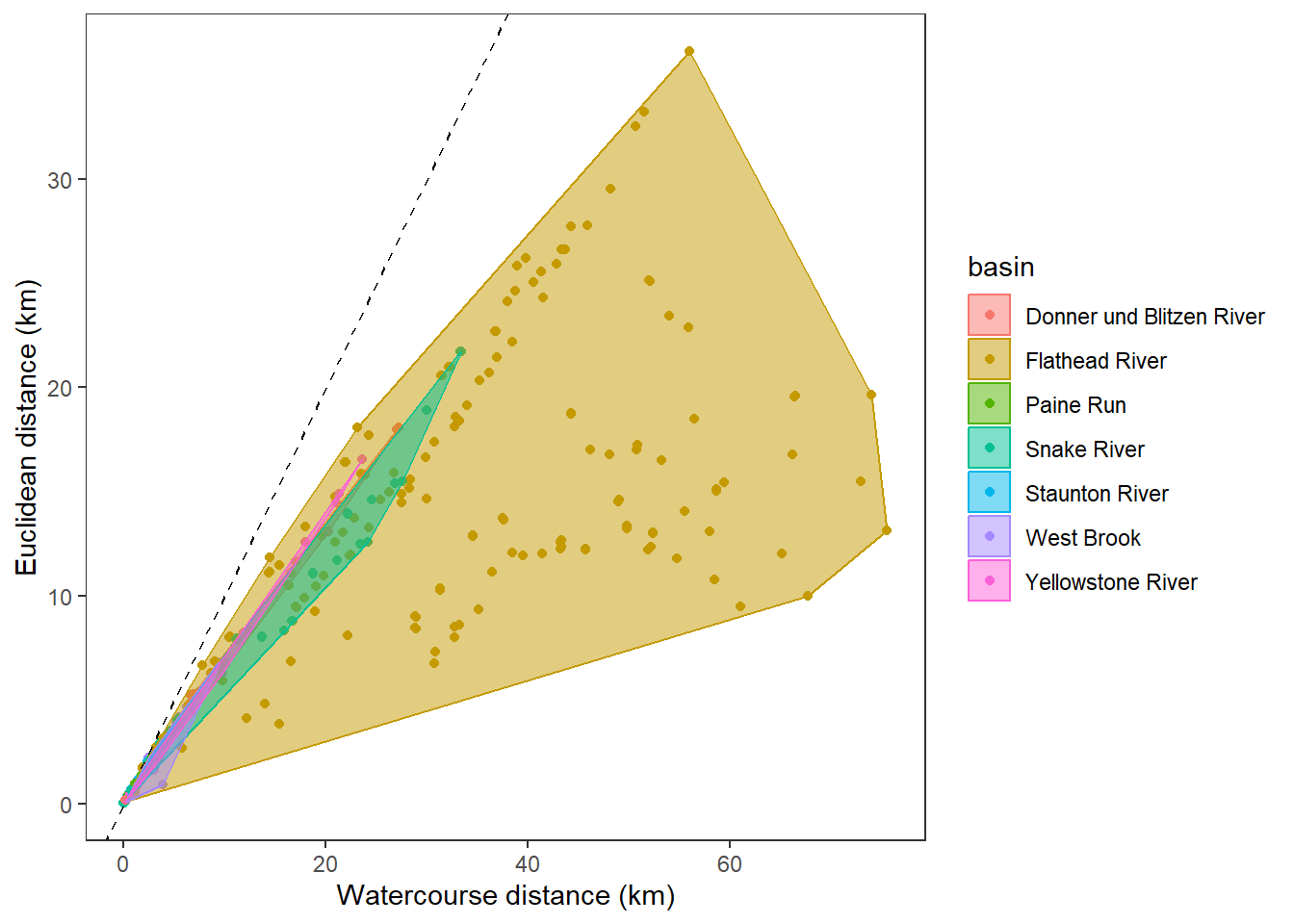
Boxplot of ratios (branching complexity)
dedw_ratio <- tibble(basin = c(rep("West Brook", times = length(as.numeric(dist_euclidean_wb / dist_watercourse_wb))),
rep("Staunton River", times = length(as.numeric(dist_euclidean_st / dist_watercourse_st))),
rep("Paine Run", times = length(as.numeric(dist_euclidean_pa / dist_watercourse_pa))),
rep("Flathead River", times = length(as.numeric(dist_euclidean_fl / dist_watercourse_fl))),
rep("Yellowstone River", times = length(as.numeric(dist_euclidean_ye / dist_watercourse_ye))),
rep("Snake River", times = length(as.numeric(dist_euclidean_sn / dist_watercourse_sn))),
rep("Donner und Blitzen River", times = length(as.numeric(dist_euclidean_db / dist_watercourse_db)))),
dedw = c(as.numeric(dist_euclidean_wb / dist_watercourse_wb),
as.numeric(dist_euclidean_st / dist_watercourse_st),
as.numeric(dist_euclidean_pa / dist_watercourse_pa),
as.numeric(dist_euclidean_fl / dist_watercourse_fl),
as.numeric(dist_euclidean_ye / dist_watercourse_ye),
as.numeric(dist_euclidean_sn / dist_watercourse_sn),
as.numeric(dist_euclidean_db / dist_watercourse_db)))
# plot
dedw_ratio %>%
mutate(basin = factor(basin, levels = c("West Brook", "Staunton River", "Paine Run", "Flathead River", "Yellowstone River", "Snake River", "Donner und Blitzen River"))) %>%
ggplot(aes(x = basin, y = dedw)) +
geom_boxplot(fill = "grey") +
ylab("Branching complexity (De/Dw)") + xlab("Basin") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.text.x = element_text(angle = 45, hjust=1))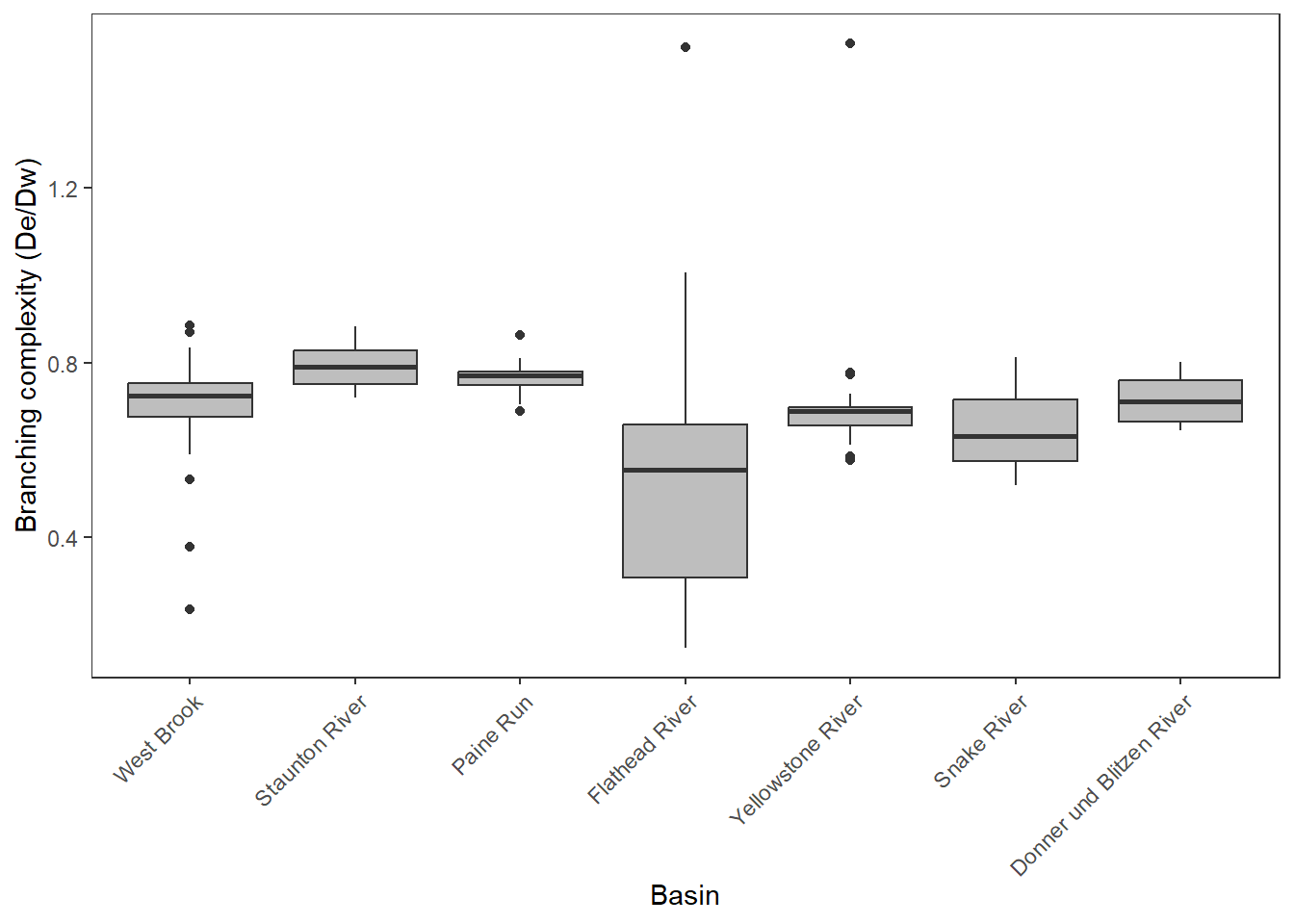
#boxplot(dedw ~ basin, dedw, ylim = c(0,1), ylab = "Branching complexity (De/Dw)", labels = c("West Brook"))Calculate Spearman’s r and time series distance
## West Brook
# basin objects
mysites <- mysites_wb
dist_euclidean <- dist_euclidean_wb
# grab
sitesss <- mysites$site_name
dat_clean2 <- dat_clean %>% filter(site_name %in% sitesss, Month %in% c(7:9))
yrs <- unique(dat_clean2$WaterYear)[c(1:4)]
# set up empty arrays
synch_arr_sp <- array(data = NA, dim = c(dim(dist_euclidean)[1], dim(dist_euclidean)[2], length(yrs)),
dimnames = list(mysites$site_name, mysites$site_name, yrs))
synch_arr_ts <- array(data = NA, dim = c(dim(dist_euclidean)[1], dim(dist_euclidean)[2], length(yrs)),
dimnames = list(mysites$site_name, mysites$site_name, yrs))
# fill arrays with Spearman's correlation coef and time series distance (euclidean)
for(k in 1:length(yrs)) {
for(i in 1:length(sitesss)) {
for(j in 1:length(sitesss)) {
dd <- dat_clean2 %>%
filter(WaterYear == yrs[k], site_name %in% c(sitesss[i], sitesss[j])) %>%
select(date, site_name, logYield) %>%
spread(key = site_name, value = logYield)
dd <- dd[complete.cases(dd),]
if(dim(dd)[1] <= 0.9*92 | dim(dd)[2] == 2) next
synch_arr_sp[i,j,k] <- cor(dd[,sitesss[i]], dd[,sitesss[j]], use = "complete.obs", method = "spearman")
synch_arr_ts[i,j,k] <- EuclideanDistance(unlist(dd[,sitesss[i]]), unlist(dd[,sitesss[j]]))
}
}
}
# set top triangles to NA
for(k in 1:length(yrs)) {
synch_arr_sp[,,k] <- ifelse(synch_arr_sp[,,k] * !upper.tri(synch_arr_sp[,,k], diag = TRUE) == 0, NA, synch_arr_sp[,,k] * !upper.tri(synch_arr_sp[,,k], diag = TRUE))
synch_arr_ts[,,k] <- ifelse(synch_arr_ts[,,k] * !upper.tri(synch_arr_ts[,,k], diag = TRUE) == 0, NA, synch_arr_ts[,,k] * !upper.tri(synch_arr_ts[,,k], diag = TRUE))
}
synch_arr_sp_wb <- synch_arr_sp
synch_arr_ts_wb <- synch_arr_ts
# Staunton
# basin objects
mysites <- mysites_st
dist_euclidean <- dist_euclidean_st
# grab
sitesss <- mysites$site_name
dat_clean2 <- dat_clean %>% filter(site_name %in% sitesss, Month %in% c(7:9))
yrs <- unique(dat_clean2$WaterYear)[c(1:3)]
# set up empty arrays
synch_arr_sp <- array(data = NA, dim = c(dim(dist_euclidean)[1], dim(dist_euclidean)[2], length(yrs)),
dimnames = list(mysites$site_name, mysites$site_name, yrs))
synch_arr_ts <- array(data = NA, dim = c(dim(dist_euclidean)[1], dim(dist_euclidean)[2], length(yrs)),
dimnames = list(mysites$site_name, mysites$site_name, yrs))
# fill arrays with Spearman's correlation coef and time series distance (euclidean)
for(k in 1:length(yrs)) {
for(i in 1:length(sitesss)) {
for(j in 1:length(sitesss)) {
dd <- dat_clean2 %>%
filter(WaterYear == yrs[k], site_name %in% c(sitesss[i], sitesss[j])) %>%
select(date, site_name, logYield) %>%
spread(key = site_name, value = logYield)
dd <- dd[complete.cases(dd),]
if(dim(dd)[1] <= 0.9*92 | dim(dd)[2] == 2) next
synch_arr_sp[i,j,k] <- cor(dd[,sitesss[i]], dd[,sitesss[j]], use = "complete.obs", method = "spearman")
synch_arr_ts[i,j,k] <- EuclideanDistance(unlist(dd[,sitesss[i]]), unlist(dd[,sitesss[j]]))
}
}
}
# set top triangles to NA
for(k in 1:length(yrs)) {
synch_arr_sp[,,k] <- ifelse(synch_arr_sp[,,k] * !upper.tri(synch_arr_sp[,,k], diag = TRUE) == 0, NA, synch_arr_sp[,,k] * !upper.tri(synch_arr_sp[,,k], diag = TRUE))
synch_arr_ts[,,k] <- ifelse(synch_arr_ts[,,k] * !upper.tri(synch_arr_ts[,,k], diag = TRUE) == 0, NA, synch_arr_ts[,,k] * !upper.tri(synch_arr_ts[,,k], diag = TRUE))
}
synch_arr_sp_st <- synch_arr_sp
synch_arr_ts_st <- synch_arr_ts
# Paine
# basin objects
mysites <- mysites_pa
dist_euclidean <- dist_euclidean_pa
# grab
sitesss <- mysites$site_name
dat_clean2 <- dat_clean %>% filter(site_name %in% sitesss, Month %in% c(7:9))
yrs <- unique(dat_clean2$WaterYear)[2]
# set up empty arrays
synch_arr_sp <- array(data = NA, dim = c(dim(dist_euclidean)[1], dim(dist_euclidean)[2], length(yrs)),
dimnames = list(mysites$site_name, mysites$site_name, yrs))
synch_arr_ts <- array(data = NA, dim = c(dim(dist_euclidean)[1], dim(dist_euclidean)[2], length(yrs)),
dimnames = list(mysites$site_name, mysites$site_name, yrs))
# fill arrays with Spearman's correlation coef and time series distance (euclidean)
for(k in 1:length(yrs)) {
for(i in 1:length(sitesss)) {
for(j in 1:length(sitesss)) {
dd <- dat_clean2 %>%
filter(WaterYear == yrs[k], site_name %in% c(sitesss[i], sitesss[j])) %>%
select(date, site_name, logYield) %>%
spread(key = site_name, value = logYield)
dd <- dd[complete.cases(dd),]
if(dim(dd)[1] <= 0.9*92 | dim(dd)[2] == 2) next
synch_arr_sp[i,j,k] <- cor(dd[,sitesss[i]], dd[,sitesss[j]], use = "complete.obs", method = "spearman")
synch_arr_ts[i,j,k] <- EuclideanDistance(unlist(dd[,sitesss[i]]), unlist(dd[,sitesss[j]]))
}
}
}
# set top triangles to NA
for(k in 1:length(yrs)) {
synch_arr_sp[,,k] <- ifelse(synch_arr_sp[,,k] * !upper.tri(synch_arr_sp[,,k], diag = TRUE) == 0, NA, synch_arr_sp[,,k] * !upper.tri(synch_arr_sp[,,k], diag = TRUE))
synch_arr_ts[,,k] <- ifelse(synch_arr_ts[,,k] * !upper.tri(synch_arr_ts[,,k], diag = TRUE) == 0, NA, synch_arr_ts[,,k] * !upper.tri(synch_arr_ts[,,k], diag = TRUE))
}
synch_arr_sp_pa <- synch_arr_sp
synch_arr_ts_pa <- synch_arr_ts
# Flathead
# basin objects
mysites <- mysites_fl
dist_euclidean <- dist_euclidean_fl
# grab
sitesss <- mysites$site_name
dat_clean2 <- dat_clean %>% filter(site_name %in% sitesss, Month %in% c(7:9))
yrs <- unique(dat_clean2$WaterYear)[c(2:5)]
# set up empty arrays
synch_arr_sp <- array(data = NA, dim = c(dim(dist_euclidean)[1], dim(dist_euclidean)[2], length(yrs)),
dimnames = list(mysites$site_name, mysites$site_name, yrs))
synch_arr_ts <- array(data = NA, dim = c(dim(dist_euclidean)[1], dim(dist_euclidean)[2], length(yrs)),
dimnames = list(mysites$site_name, mysites$site_name, yrs))
# fill arrays with Spearman's correlation coef and time series distance (euclidean)
for(k in 1:length(yrs)) {
for(i in 1:length(sitesss)) {
for(j in 1:length(sitesss)) {
dd <- dat_clean2 %>%
filter(WaterYear == yrs[k], site_name %in% c(sitesss[i], sitesss[j])) %>%
select(date, site_name, logYield) %>%
spread(key = site_name, value = logYield)
dd <- dd[complete.cases(dd),]
if(dim(dd)[1] <= 0.9*92 | dim(dd)[2] == 2) next
synch_arr_sp[i,j,k] <- cor(dd[,sitesss[i]], dd[,sitesss[j]], use = "complete.obs", method = "spearman")
synch_arr_ts[i,j,k] <- EuclideanDistance(unlist(dd[,sitesss[i]]), unlist(dd[,sitesss[j]]))
}
}
}
# set top triangles to NA
for(k in 1:length(yrs)) {
synch_arr_sp[,,k] <- ifelse(synch_arr_sp[,,k] * !upper.tri(synch_arr_sp[,,k], diag = TRUE) == 0, NA, synch_arr_sp[,,k] * !upper.tri(synch_arr_sp[,,k], diag = TRUE))
synch_arr_ts[,,k] <- ifelse(synch_arr_ts[,,k] * !upper.tri(synch_arr_ts[,,k], diag = TRUE) == 0, NA, synch_arr_ts[,,k] * !upper.tri(synch_arr_ts[,,k], diag = TRUE))
}
synch_arr_sp_fl <- synch_arr_sp
synch_arr_ts_fl <- synch_arr_ts
# Yellowstone
# basin objects
mysites <- mysites_ye
dist_euclidean <- dist_euclidean_ye
# grab
sitesss <- mysites$site_name
dat_clean2 <- dat_clean %>% filter(site_name %in% sitesss, Month %in% c(7:9))
yrs <- unique(dat_clean2$WaterYear)[c(2,3,5)]
# set up empty arrays
synch_arr_sp <- array(data = NA, dim = c(dim(dist_euclidean)[1], dim(dist_euclidean)[2], length(yrs)),
dimnames = list(mysites$site_name, mysites$site_name, yrs))
synch_arr_ts <- array(data = NA, dim = c(dim(dist_euclidean)[1], dim(dist_euclidean)[2], length(yrs)),
dimnames = list(mysites$site_name, mysites$site_name, yrs))
# fill arrays with Spearman's correlation coef and time series distance (euclidean)
for(k in 1:length(yrs)) {
for(i in 1:length(sitesss)) {
for(j in 1:length(sitesss)) {
dd <- dat_clean2 %>%
filter(WaterYear == yrs[k], site_name %in% c(sitesss[i], sitesss[j])) %>%
select(date, site_name, logYield) %>%
spread(key = site_name, value = logYield)
dd <- dd[complete.cases(dd),]
if(dim(dd)[1] <= 0.9*92 | dim(dd)[2] == 2) next
synch_arr_sp[i,j,k] <- cor(dd[,sitesss[i]], dd[,sitesss[j]], use = "complete.obs", method = "spearman")
synch_arr_ts[i,j,k] <- EuclideanDistance(unlist(dd[,sitesss[i]]), unlist(dd[,sitesss[j]]))
}
}
}
# set top triangles to NA
for(k in 1:length(yrs)) {
synch_arr_sp[,,k] <- ifelse(synch_arr_sp[,,k] * !upper.tri(synch_arr_sp[,,k], diag = TRUE) == 0, NA, synch_arr_sp[,,k] * !upper.tri(synch_arr_sp[,,k], diag = TRUE))
synch_arr_ts[,,k] <- ifelse(synch_arr_ts[,,k] * !upper.tri(synch_arr_ts[,,k], diag = TRUE) == 0, NA, synch_arr_ts[,,k] * !upper.tri(synch_arr_ts[,,k], diag = TRUE))
}
synch_arr_sp_ye <- synch_arr_sp
synch_arr_ts_ye <- synch_arr_ts
# Snake
# basin objects
mysites <- mysites_sn
dist_euclidean <- dist_euclidean_sn
# grab
sitesss <- mysites$site_name
dat_clean2 <- dat_clean %>% filter(site_name %in% sitesss, Month %in% c(7:9))
yrs <- unique(dat_clean2$WaterYear)[c(1,3,4)]
# set up empty arrays
synch_arr_sp <- array(data = NA, dim = c(dim(dist_euclidean)[1], dim(dist_euclidean)[2], length(yrs)),
dimnames = list(mysites$site_name, mysites$site_name, yrs))
synch_arr_ts <- array(data = NA, dim = c(dim(dist_euclidean)[1], dim(dist_euclidean)[2], length(yrs)),
dimnames = list(mysites$site_name, mysites$site_name, yrs))
# fill arrays with Spearman's correlation coef and time series distance (euclidean)
for(k in 1:length(yrs)) {
for(i in 1:length(sitesss)) {
for(j in 1:length(sitesss)) {
dd <- dat_clean2 %>%
filter(WaterYear == yrs[k], site_name %in% c(sitesss[i], sitesss[j])) %>%
select(date, site_name, logYield) %>%
spread(key = site_name, value = logYield)
dd <- dd[complete.cases(dd),]
if(dim(dd)[1] <= 0.9*92 | dim(dd)[2] == 2) next
synch_arr_sp[i,j,k] <- cor(dd[,sitesss[i]], dd[,sitesss[j]], use = "complete.obs", method = "spearman")
synch_arr_ts[i,j,k] <- EuclideanDistance(unlist(dd[,sitesss[i]]), unlist(dd[,sitesss[j]]))
}
}
}
# set top triangles to NA
for(k in 1:length(yrs)) {
synch_arr_sp[,,k] <- ifelse(synch_arr_sp[,,k] * !upper.tri(synch_arr_sp[,,k], diag = TRUE) == 0, NA, synch_arr_sp[,,k] * !upper.tri(synch_arr_sp[,,k], diag = TRUE))
synch_arr_ts[,,k] <- ifelse(synch_arr_ts[,,k] * !upper.tri(synch_arr_ts[,,k], diag = TRUE) == 0, NA, synch_arr_ts[,,k] * !upper.tri(synch_arr_ts[,,k], diag = TRUE))
}
synch_arr_sp_sn <- synch_arr_sp
synch_arr_ts_sn <- synch_arr_ts
# Donner Blitzen
# basin objects
mysites <- mysites_db
dist_euclidean <- dist_euclidean_db
# grab
sitesss <- mysites$site_name
dat_clean2 <- dat_clean %>% filter(site_name %in% sitesss, Month %in% c(7:9))
yrs <- unique(dat_clean2$WaterYear)[c(2:4)]
# set up empty arrays
synch_arr_sp <- array(data = NA, dim = c(dim(dist_euclidean)[1], dim(dist_euclidean)[2], length(yrs)),
dimnames = list(mysites$site_name, mysites$site_name, yrs))
synch_arr_ts <- array(data = NA, dim = c(dim(dist_euclidean)[1], dim(dist_euclidean)[2], length(yrs)),
dimnames = list(mysites$site_name, mysites$site_name, yrs))
# fill arrays with Spearman's correlation coef and time series distance (euclidean)
for(k in 1:length(yrs)) {
for(i in 1:length(sitesss)) {
for(j in 1:length(sitesss)) {
dd <- dat_clean2 %>%
filter(WaterYear == yrs[k], site_name %in% c(sitesss[i], sitesss[j])) %>%
select(date, site_name, logYield) %>%
spread(key = site_name, value = logYield)
dd <- dd[complete.cases(dd),]
if(dim(dd)[1] <= 0.9*92 | dim(dd)[2] == 2) next
synch_arr_sp[i,j,k] <- cor(dd[,sitesss[i]], dd[,sitesss[j]], use = "complete.obs", method = "spearman")
synch_arr_ts[i,j,k] <- EuclideanDistance(unlist(dd[,sitesss[i]]), unlist(dd[,sitesss[j]]))
}
}
}
# set top triangles to NA
for(k in 1:length(yrs)) {
synch_arr_sp[,,k] <- ifelse(synch_arr_sp[,,k] * !upper.tri(synch_arr_sp[,,k], diag = TRUE) == 0, NA, synch_arr_sp[,,k] * !upper.tri(synch_arr_sp[,,k], diag = TRUE))
synch_arr_ts[,,k] <- ifelse(synch_arr_ts[,,k] * !upper.tri(synch_arr_ts[,,k], diag = TRUE) == 0, NA, synch_arr_ts[,,k] * !upper.tri(synch_arr_ts[,,k], diag = TRUE))
}
synch_arr_sp_db <- synch_arr_sp
synch_arr_ts_db <- synch_arr_tsCreate plotting function
fluvsynchfun <- function(subbas, spear, ts, dist_euc, dist_wat, dist_flow) {
synch_arr_sp <- spear
synch_arr_ts <- ts
dist_euclidean <- dist_euc
dist_watercourse <- dist_wat
dist_flowconn <- dist_flow
synchtib <- tibble(basin = rep(subbas, times = length(as.numeric(synch_arr_sp))),
wateryear = sort(rep(as.numeric(dimnames(synch_arr_sp)[[3]]), times = dim(synch_arr_sp)[1]*dim(synch_arr_sp)[2])),
synch_sp = as.numeric(synch_arr_sp),
synch_ts = as.numeric(synch_arr_ts),
euclidean = rep(as.numeric(dist_euclidean), times = dim(synch_arr_sp)[3]),
watercourse = rep(as.numeric(dist_watercourse), times = dim(synch_arr_sp)[3]),
flowconnected = rep(as.numeric(dist_flowconn), times = dim(synch_arr_sp)[3])) %>%
gather(euclidean:flowconnected, key = "distance", value = "value")
# Spearman correlation
fs_sp <- synchtib %>%
mutate(distance = factor(distance, levels = c("flowconnected", "watercourse", "euclidean"))) %>%
ggplot(aes(x = value, y = synch_sp, color = distance, fill = distance, group = distance)) +
geom_smooth(alpha = 0.2, se = FALSE, span = 1) +
geom_point(alpha = 0.5) +
scale_color_manual(values = c("blue", "purple", "gold1")) +
scale_fill_manual(values = c("blue", "purple", "gold1")) +
facet_wrap(~wateryear, nrow = 1) +
ylab("Spearman's corr") + xlab("Distance (km)") +
#coord_cartesian(ylim = c(0,1)) +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), axis.title.x=element_blank())
# Time series distance (euclidean)
fs_ts <- synchtib %>%
mutate(distance = factor(distance, levels = c("flowconnected", "watercourse", "euclidean"))) %>%
ggplot(aes(x = value, y = synch_ts, color = distance, fill = distance, group = distance)) +
geom_smooth(alpha = 0.2, se = FALSE, span = 1) +
geom_point(alpha = 0.5) +
scale_color_manual(values = c("blue", "purple", "gold1")) +
scale_fill_manual(values = c("blue", "purple", "gold1")) +
facet_wrap(~wateryear, nrow = 1) +
ylab("TS distance") + xlab("Distance (km)") +
#coord_cartesian(ylim = c(0,20)) +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), axis.title.x=element_blank())
#
# # Spearman correlation
# fs_sp2 <- synchtib %>%
# filter(distance %in% c("dist_flo")) %>%
# mutate(distance = factor(distance, levels = c("dist_flo", "dist_dis"))) %>%
# ggplot(aes(x = value, y = synch_sp, color = distance, fill = distance, group = distance)) +
# geom_smooth(alpha = 0.2) +
# geom_point(alpha = 0.5) +
# scale_color_manual(values = c("blue", "darkorange")) +
# scale_fill_manual(values = c("blue", "darkorange")) +
# facet_wrap(~wateryear, nrow = 1) +
# ylab("Spearman's corr.") + xlab("Distance (km)") +
# coord_cartesian(ylim = c(0,1)) +
# theme_bw() +
# theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), axis.title.x=element_blank())
#
# # Time series distance (euclidean)
# fs_ts2 <- synchtib %>%
# filter(distance %in% c("dist_flo", "dist_dis")) %>%
# mutate(distance = factor(distance, levels = c("dist_flo", "dist_dis"))) %>%
# ggplot(aes(x = value, y = synch_ts, color = distance, fill = distance, group = distance)) +
# geom_smooth(alpha = 0.2) +
# geom_point(alpha = 0.5) +
# scale_color_manual(values = c("blue", "darkorange")) +
# scale_fill_manual(values = c("blue", "darkorange")) +
# facet_wrap(~wateryear, nrow = 1) +
# ylab("TS distance") + xlab("Distance (km)") +
# coord_cartesian(ylim = c(0,20)) +
# theme_bw() +
# theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), axis.title.x=element_blank())
#
myplot <- ggarrange(fs_sp, fs_ts, ncol = 1)
annotate_figure(myplot, bottom = text_grob("Distance (km)"))
}Plot fluvial synchrograms
fluvsynchfun(subbas = "West Brook", spear = synch_arr_sp_wb, ts = synch_arr_ts_wb,
dist_euc = dist_euclidean_wb, dist_wat = dist_watercourse_wb, dist_flow = dist_flowconn_wb)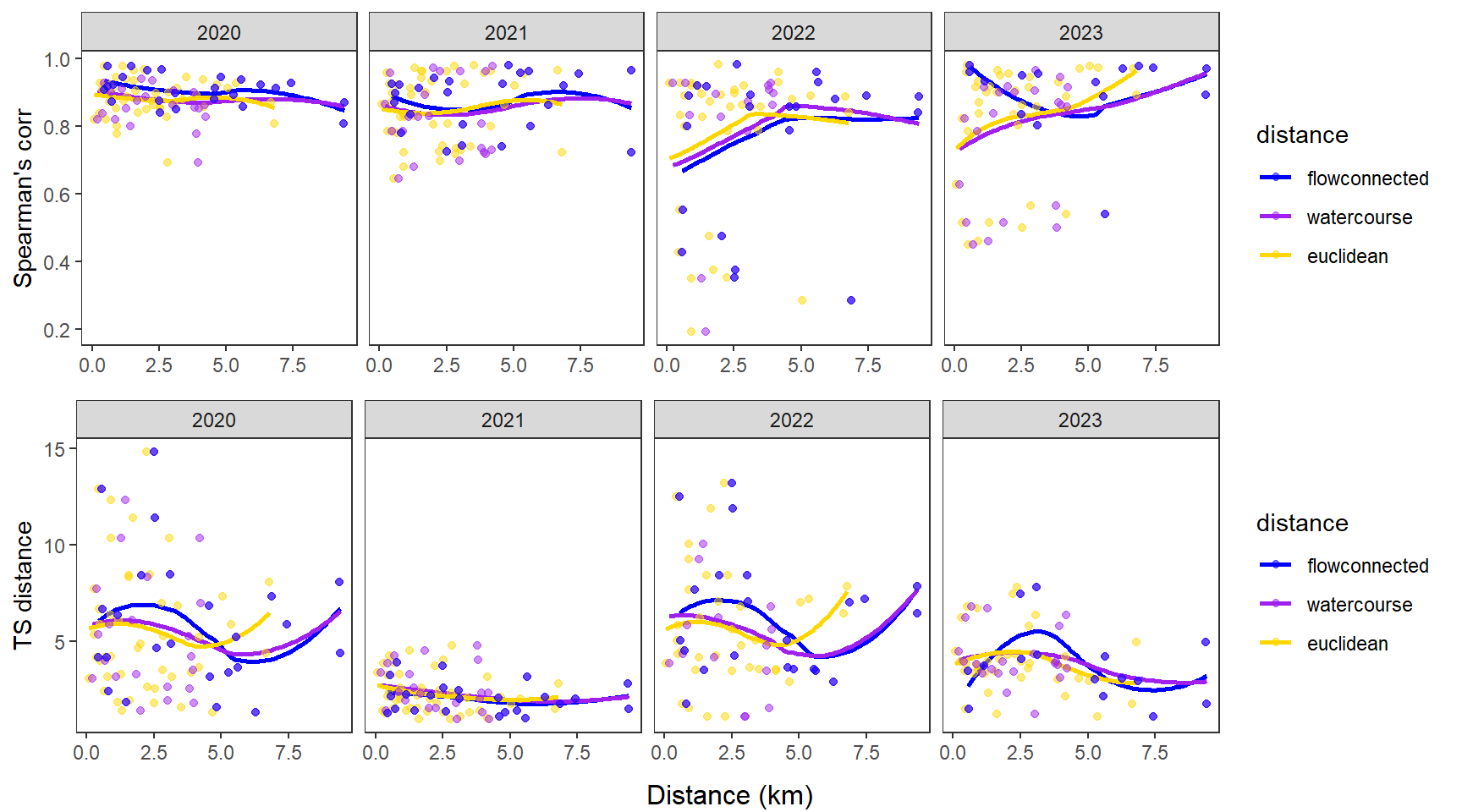
fluvsynchfun(subbas = "Staunton River", spear = synch_arr_sp_st, ts = synch_arr_ts_st,
dist_euc = dist_euclidean_st, dist_wat = dist_watercourse_st, dist_flow = dist_flowconn_st)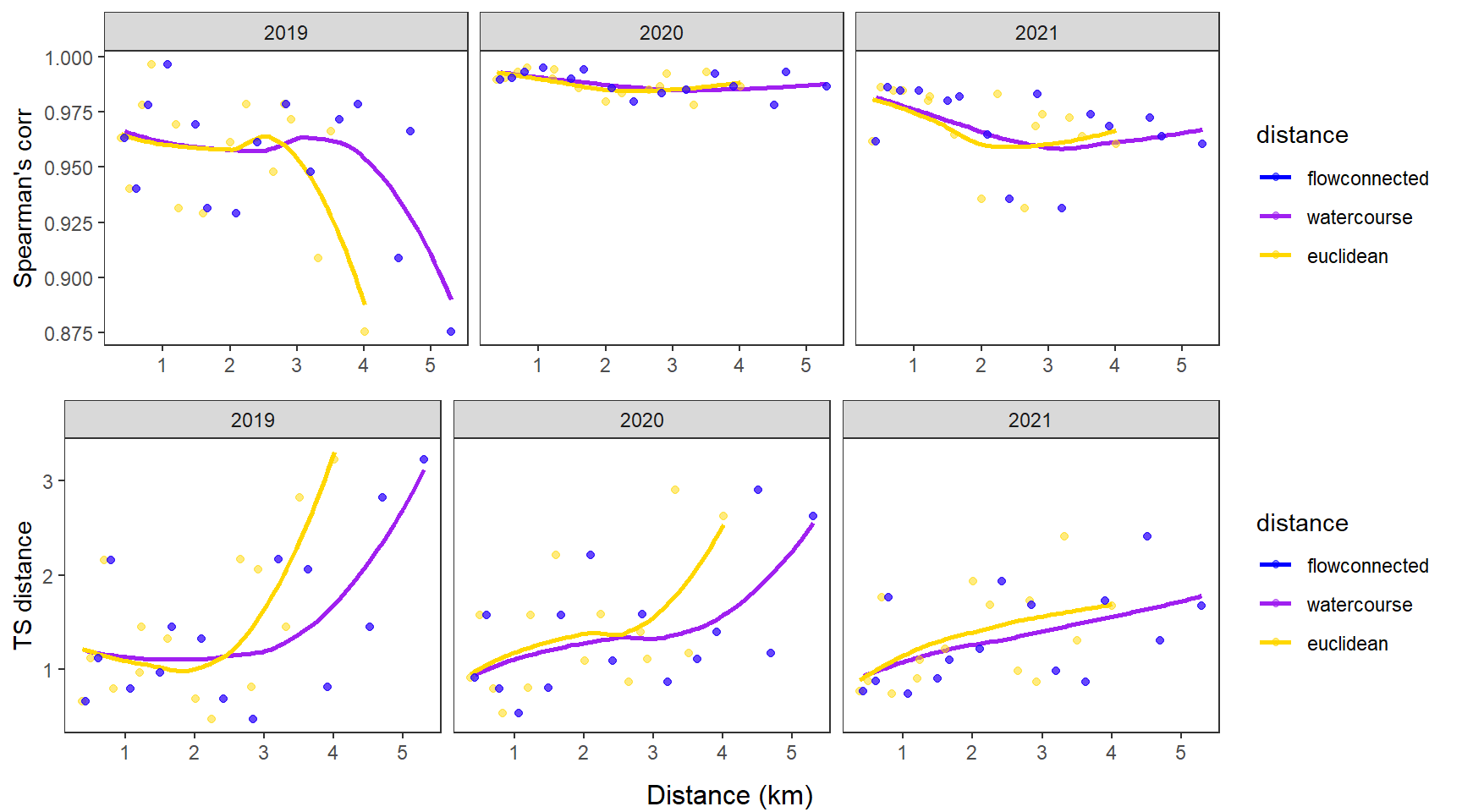
fluvsynchfun(subbas = "Paine Run", spear = synch_arr_sp_pa, ts = synch_arr_ts_pa,
dist_euc = dist_euclidean_pa, dist_wat = dist_watercourse_pa, dist_flow = dist_flowconn_pa)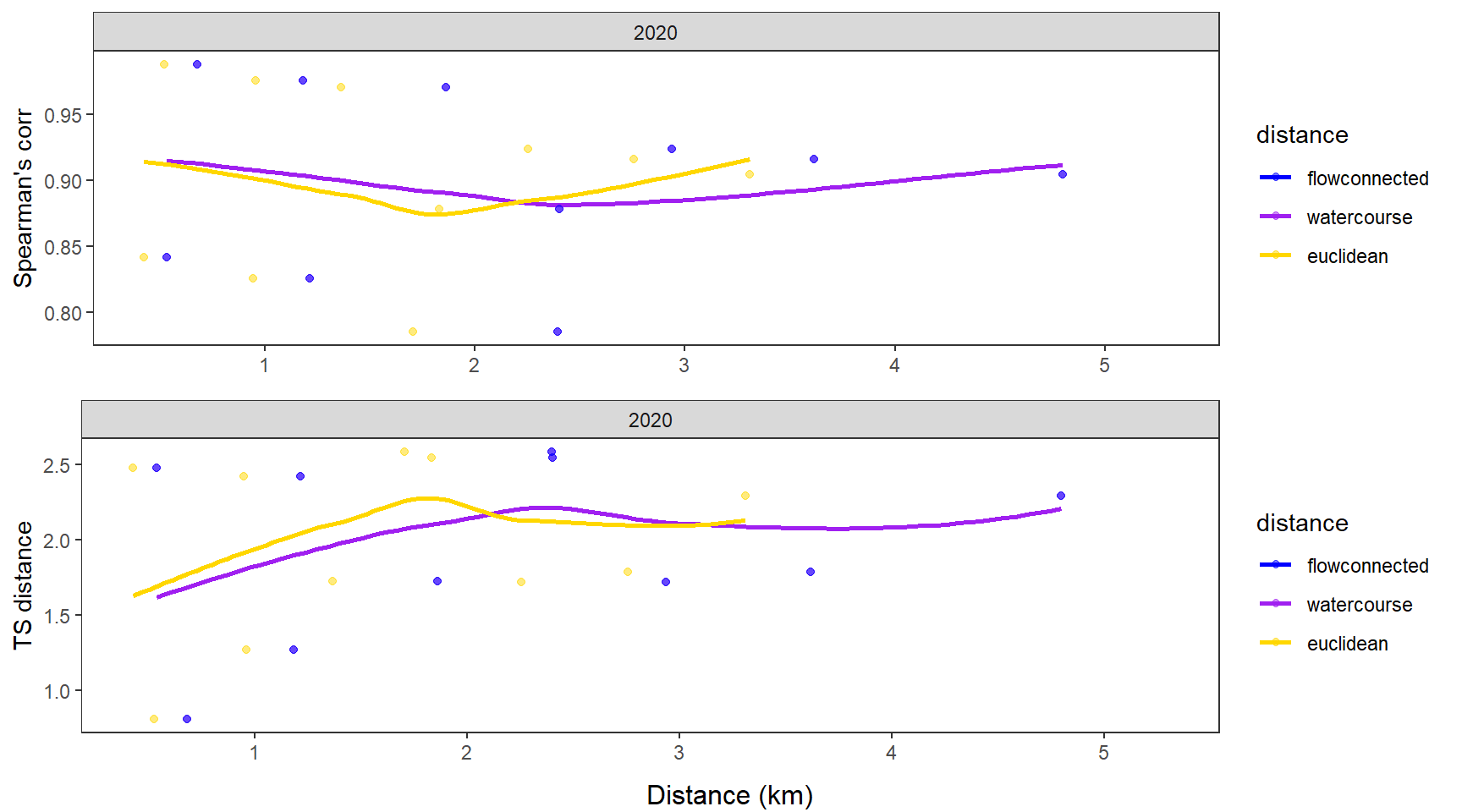
fluvsynchfun(subbas = "Flathead River", spear = synch_arr_sp_fl, ts = synch_arr_ts_fl,
dist_euc = dist_euclidean_fl, dist_wat = dist_watercourse_fl, dist_flow = dist_flowconn_fl)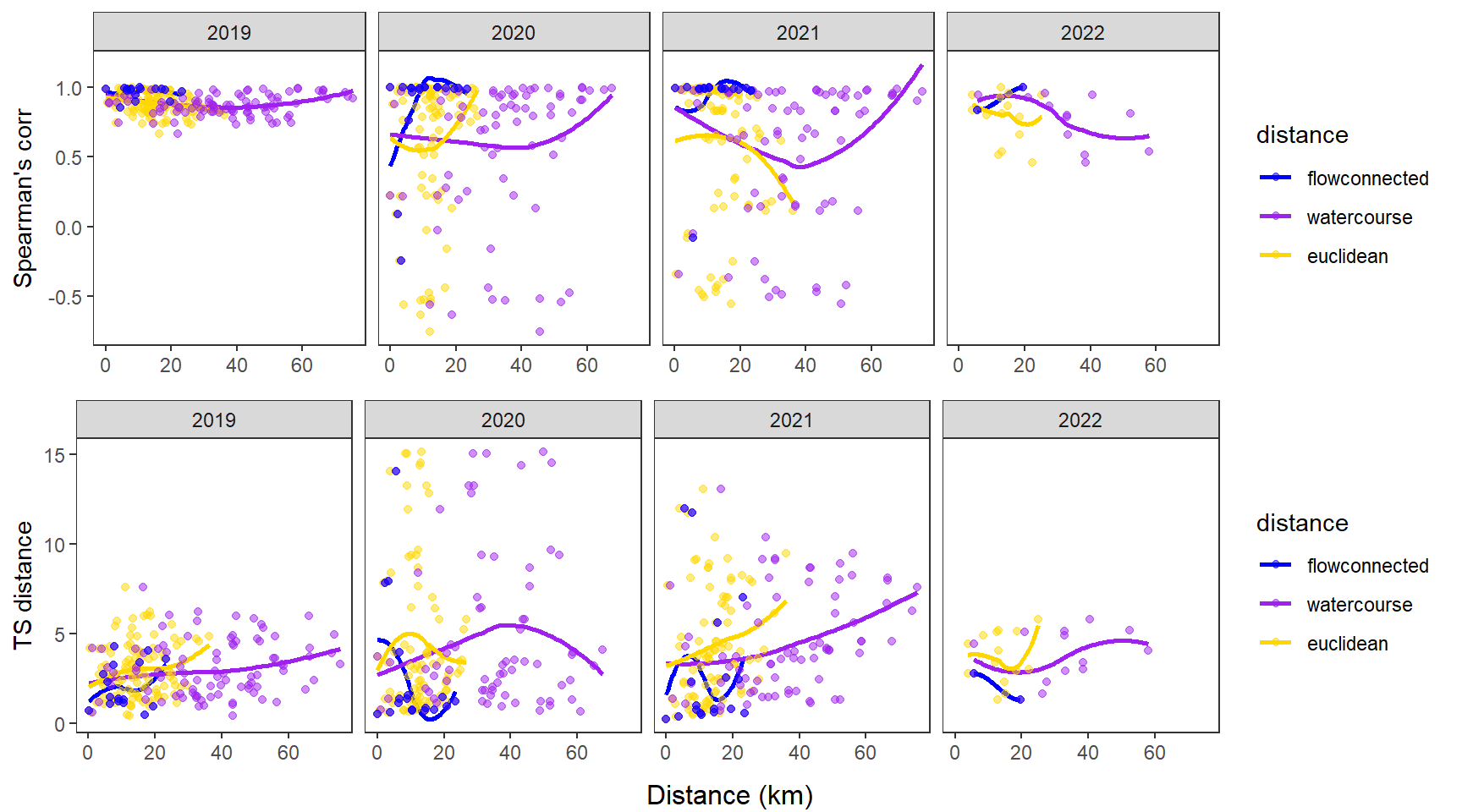
fluvsynchfun(subbas = "Yellowstone River", spear = synch_arr_sp_ye, ts = synch_arr_ts_ye,
dist_euc = dist_euclidean_ye, dist_wat = dist_watercourse_ye, dist_flow = dist_flowconn_ye)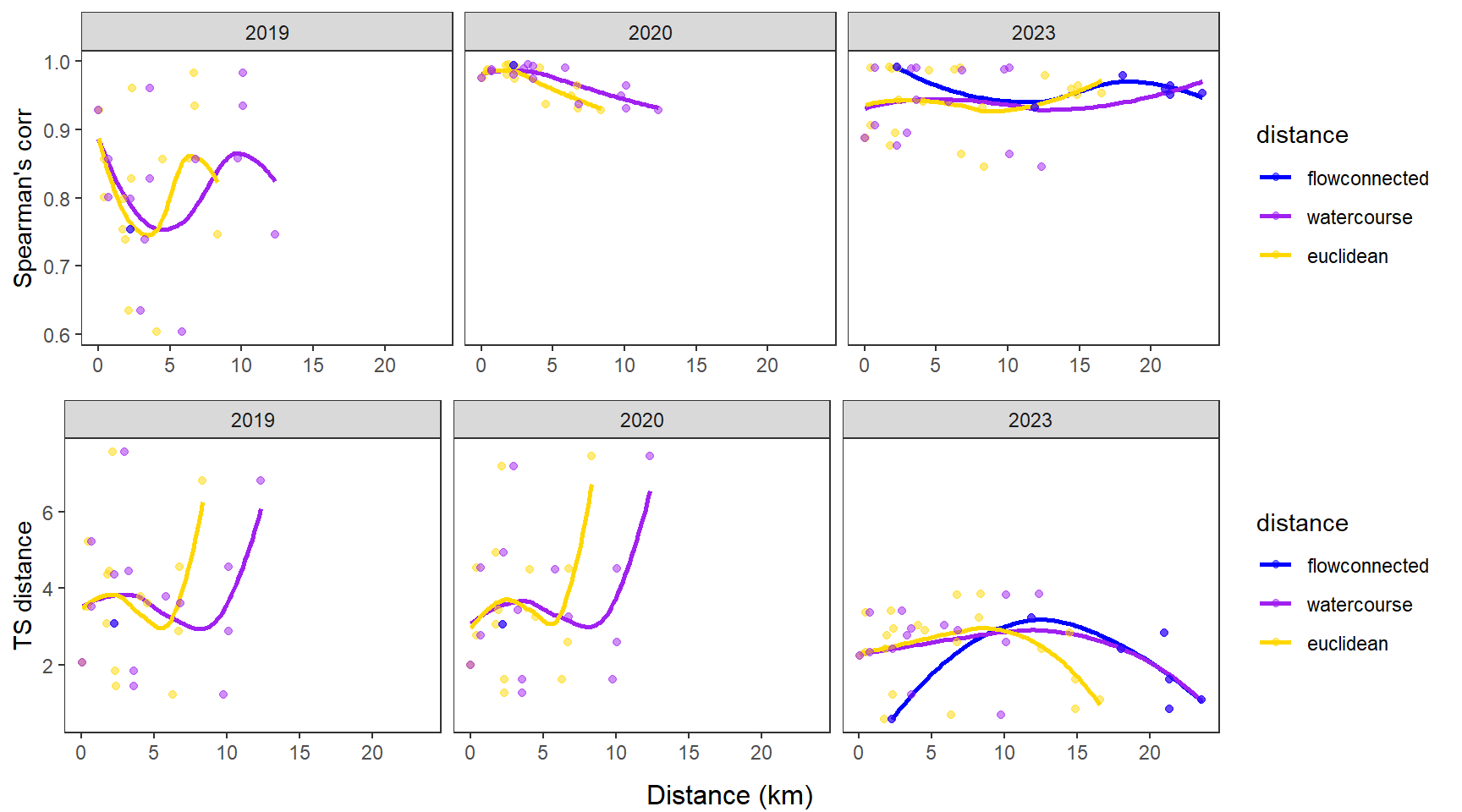
fluvsynchfun(subbas = "Snake River", spear = synch_arr_sp_sn, ts = synch_arr_ts_sn,
dist_euc = dist_euclidean_sn, dist_wat = dist_watercourse_sn, dist_flow = dist_flowconn_sn)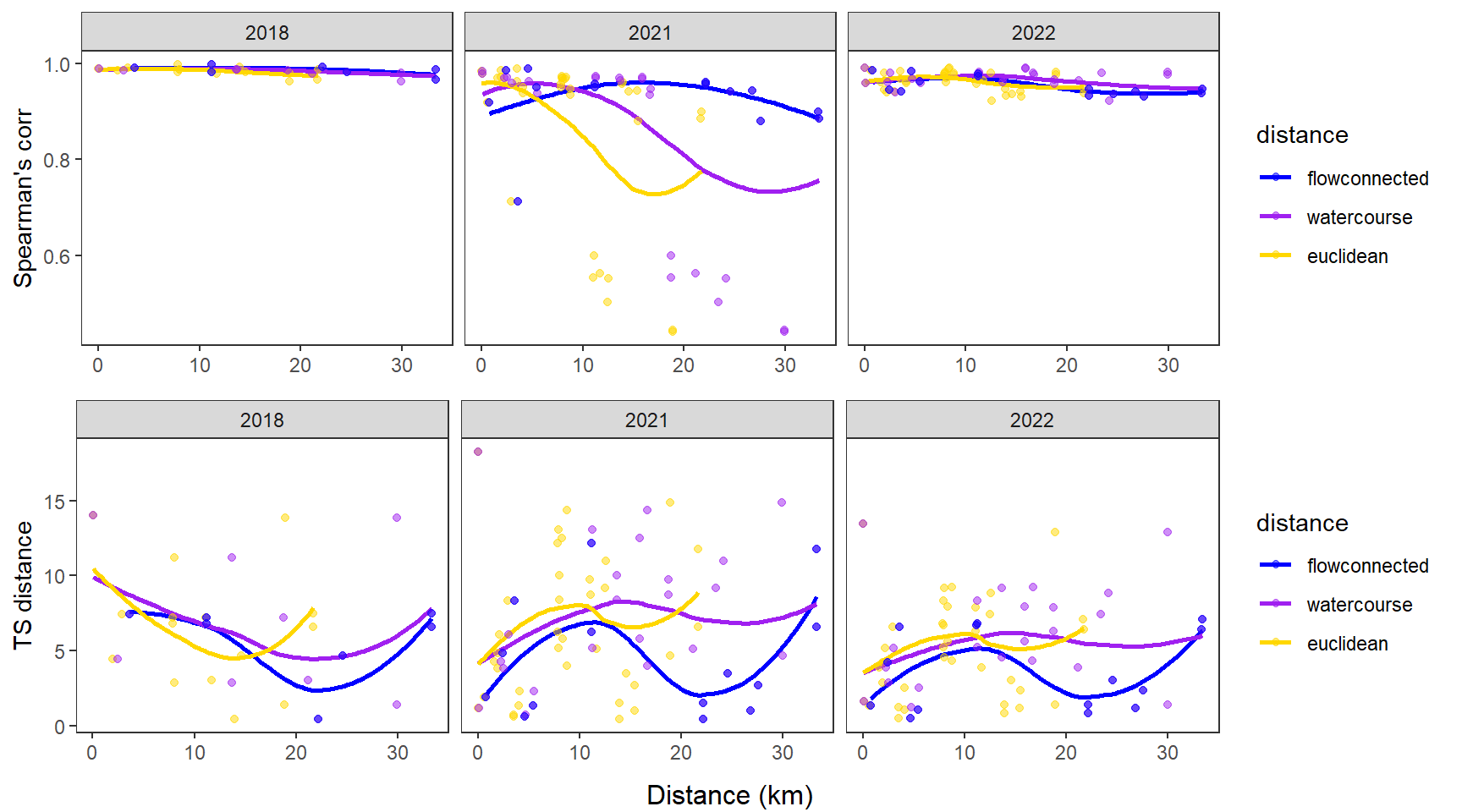
fluvsynchfun(subbas = "Donner und Blitzen River", spear = synch_arr_sp_db, ts = synch_arr_ts_db,
dist_euc = dist_euclidean_db, dist_wat = dist_watercourse_db, dist_flow = dist_flowconn_db)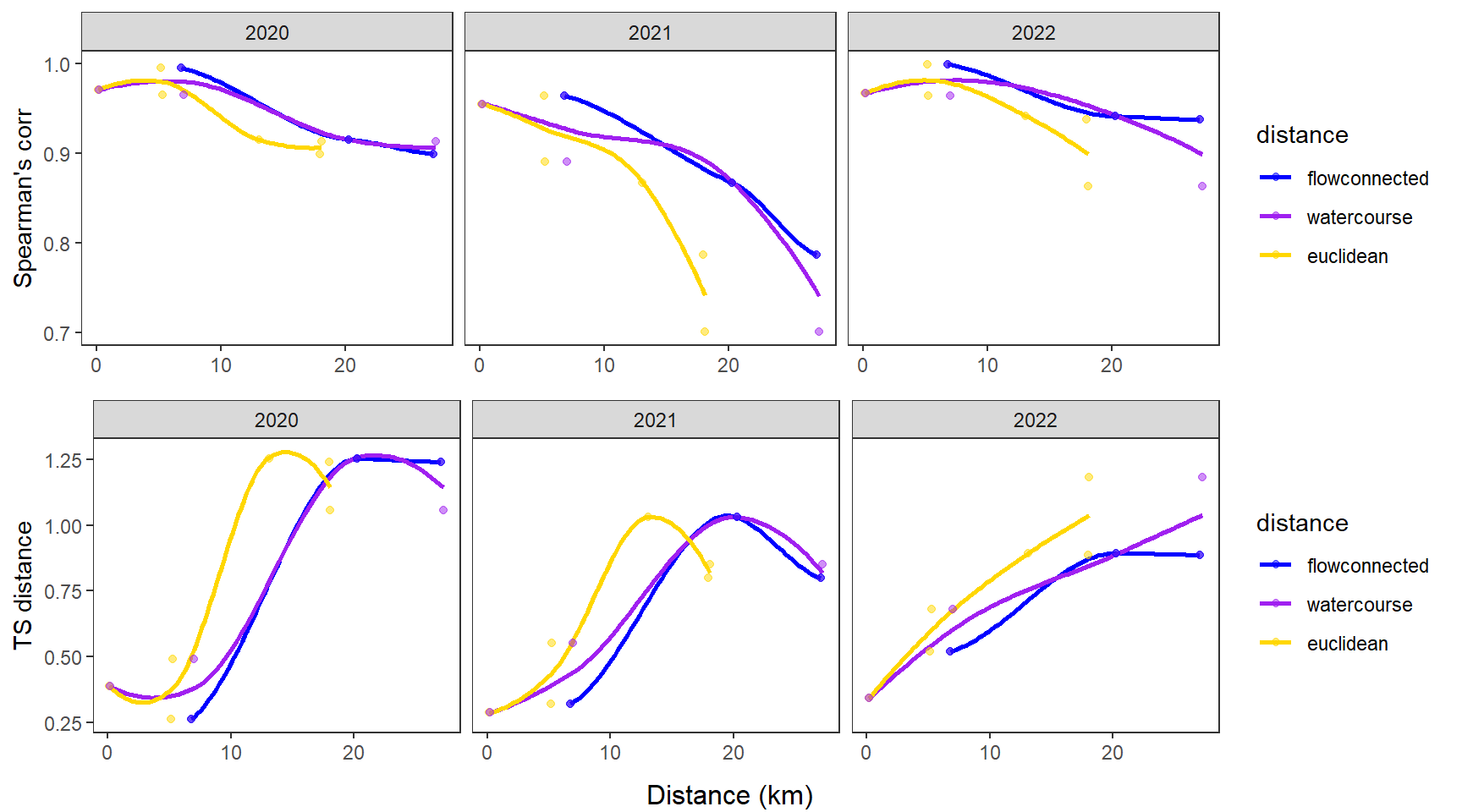
synchtib <- tibble(basin = c(rep("West Brook", times = length(as.numeric(synch_mat))),
rep("Snake River", times = length(as.numeric(synch_mat_snake)))),
synch_sp = c(as.numeric(synch_mat_sp), as.numeric(synch_mat_snake_sp)),
synch_ts = c(as.numeric(synch_mat_ts), as.numeric(synch_mat_snake_ts)),
dist_euc = c(as.numeric(dist_euclidean), as.numeric(dist_euclidean_snake)),
dist_wat = c(as.numeric(dist_watercourse), as.numeric(dist_watercourse_snake)),
dist_flo = c(as.numeric(dist_flowconn), as.numeric(dist_flowconn_snake)),
dist_dis = c(as.numeric(dist_flowdisconn), as.numeric(dist_flowdisconn_snake))) %>%
gather(dist_euc:dist_dis, key = "distance", value = "value")
# Spearman correlation
synchtib %>%
filter(!distance %in% c("dist_dis")) %>%
mutate(distance = factor(distance, levels = c("dist_flo", "dist_wat", "dist_euc"))) %>%
ggplot(aes(x = value, y = synch_sp, color = distance, fill = distance, group = distance)) +
geom_smooth(alpha = 0.3) +
geom_point(aes(x = jitter(value, factor = 300), y = jitter(synch_sp, factor = 300))) +
scale_color_manual(values = c("blue", "purple", "gold1")) +
scale_fill_manual(values = c("blue", "purple", "gold1")) +
facet_wrap(~basin, scales = "free_x") +
ylab("Spearman's correlation") + xlab("Distance (km)") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
# Time series distance (euclidean)
synchtib %>%
filter(!distance %in% c("dist_dis")) %>%
mutate(distance = factor(distance, levels = c("dist_flo", "dist_wat", "dist_euc")))%>%
ggplot(aes(x = value, y = synch_ts, color = distance, fill = distance, group = distance)) +
geom_smooth(alpha = 0.3) +
geom_point(aes(x = jitter(value, factor = 200), y = jitter(synch_ts, factor = 200))) +
scale_color_manual(values = c("blue", "purple", "gold1")) +
scale_fill_manual(values = c("blue", "purple", "gold1")) +
facet_wrap(~basin, scales = "free_x") +
ylab("Time series distance") + xlab("Distance (km)") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
# Spearman correlation
synchtib %>%
filter(distance %in% c("dist_flo", "dist_dis")) %>%
mutate(distance = factor(distance, levels = c("dist_flo", "dist_dis"))) %>%
ggplot(aes(x = value, y = synch_sp, color = distance, fill = distance, group = distance)) +
geom_smooth(alpha = 0.3) +
geom_point(aes(x = jitter(value, factor = 300), y = jitter(synch_sp, factor = 300))) +
scale_color_manual(values = c("blue", "darkorange")) +
scale_fill_manual(values = c("blue", "darkorange")) +
facet_wrap(~basin, scales = "free_x") +
ylab("Spearman's correlation") + xlab("Distance (km)") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
# Time series distance (euclidean)
synchtib %>%
filter(distance %in% c("dist_flo", "dist_dis")) %>%
mutate(distance = factor(distance, levels = c("dist_flo", "dist_dis"))) %>%
ggplot(aes(x = value, y = synch_ts, color = distance, fill = distance, group = distance)) +
geom_smooth(alpha = 0.3) +
geom_point(aes(x = jitter(value, factor = 200), y = jitter(synch_ts, factor = 200))) +
scale_color_manual(values = c("blue", "darkorange")) +
scale_fill_manual(values = c("blue", "darkorange")) +
facet_wrap(~basin, scales = "free_x") +
ylab("Time series distance") + xlab("Distance (km)") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())tibble(basin = c(rep("West Brook", times = length(as.numeric(synch_mat))),
rep("Snake River", times = length(as.numeric(synch_mat_snake)))),
synch_sp = c(as.numeric(synch_mat_sp), as.numeric(synch_mat_snake_sp)),
synch_ts = c(as.numeric(synch_mat_ts), as.numeric(synch_mat_snake_ts)),
dist_euc = c(as.numeric(dist_euclidean), as.numeric(dist_euclidean_snake)),
dist_wat = c(as.numeric(dist_watercourse), as.numeric(dist_watercourse_snake)),
dist_flo = c(as.numeric(dist_flowconn), as.numeric(dist_flowconn_snake)),
dist_dis = c(as.numeric(dist_flowdisconn), as.numeric(dist_flowdisconn_snake))) %>%
mutate(dedw = dist_euc / dist_wat) %>%
ggplot(aes(x = dedw, y = synch_ts)) +
geom_smooth(alpha = 0.3, method = "lm") +
geom_point(aes(x = dedw, y = synch_ts)) +
facet_wrap(~basin, scales = "free_x") +
ylab("Time series distance") + xlab("de/dw") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
tibble(basin = c(rep("West Brook", times = length(as.numeric(synch_mat))),
rep("Snake River", times = length(as.numeric(synch_mat_snake)))),
synch_sp = c(as.numeric(synch_mat_sp), as.numeric(synch_mat_snake_sp)),
synch_ts = c(as.numeric(synch_mat_ts), as.numeric(synch_mat_snake_ts)),
dist_euc = c(as.numeric(dist_euclidean), as.numeric(dist_euclidean_snake)),
dist_wat = c(as.numeric(dist_watercourse), as.numeric(dist_watercourse_snake)),
dist_flo = c(as.numeric(dist_flowconn), as.numeric(dist_flowconn_snake)),
dist_dis = c(as.numeric(dist_flowdisconn), as.numeric(dist_flowdisconn_snake))) %>%
mutate(dedw = dist_euc / dist_wat) %>%
ggplot(aes(x = dedw, y = synch_sp)) +
geom_smooth(alpha = 0.3, method = "lm") +
geom_point(aes(x = dedw, y = synch_sp)) +
facet_wrap(~basin, scales = "free_x") +
ylab("Spearman correlation") + xlab("de/dw") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())Plotting function
fluvsynchfun_mod <- function(subbas, spear, ts, dist_euc, dist_wat, dist_flow) {
synch_arr_sp <- spear
synch_arr_ts <- ts
dist_euclidean <- dist_euc
dist_watercourse <- dist_wat
dist_flowconn <- dist_flow
synchtib <- tibble(basin = rep(subbas, times = length(as.numeric(synch_arr_sp))),
wateryear = sort(rep(as.numeric(dimnames(synch_arr_sp)[[3]]), times = dim(synch_arr_sp)[1]*dim(synch_arr_sp)[2])),
synch_sp = as.numeric(synch_arr_sp),
synch_ts = as.numeric(synch_arr_ts),
euclidean = rep(as.numeric(dist_euclidean), times = dim(synch_arr_sp)[3]),
watercourse = rep(as.numeric(dist_watercourse), times = dim(synch_arr_sp)[3]),
flowconnected = rep(as.numeric(dist_flowconn), times = dim(synch_arr_sp)[3])) %>%
gather(euclidean:flowconnected, key = "distance", value = "value")
# Spearman correlation
# fs_sp <- synchtib %>%
# filter(distance != "watercourse") %>%
# mutate(distance = factor(distance, levels = c("flowconnected", "watercourse", "euclidean"))) %>%
# ggplot(aes(x = value, y = synch_sp, color = distance, fill = distance, group = distance)) +
# geom_smooth(alpha = 0.2, se = FALSE) +
# geom_point(alpha = 0.5) +
# scale_color_manual(values = c("blue", "purple")) +
# scale_fill_manual(values = c("blue", "purple")) +
# facet_wrap(~wateryear, nrow = 1) +
# ylab("Spearman's corr") + xlab("Distance (km)") +
# #coord_cartesian(ylim = c(0,1)) +
# theme_bw() +
# theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), axis.title.x=element_blank())
# Time series distance (euclidean)
fs_ts <- synchtib %>%
filter(distance != "watercourse") %>%
mutate(distance = factor(distance, levels = c("euclidean", "flowconnected"))) %>%
ggplot(aes(x = value, y = synch_ts, color = distance, fill = distance, group = distance)) +
geom_smooth(alpha = 0.2, span = 1, span = 1) +
geom_point(alpha = 0.2) +
scale_color_manual(values = c("blue", "darkorange")) +
scale_fill_manual(values = c("blue", "darkorange")) +
ggh4x::facet_wrap2(~wateryear, nrow = 1, ncol = 4, trim_blank = FALSE) +
ylab("") + xlab("") + ggtitle(subbas) +
#coord_cartesian(ylim = c(0,20)) +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.title.x=element_blank(), axis.title.y = element_blank())
# myplot <- ggarrange(fs_sp, fs_ts, ncol = 1)
# annotate_figure(myplot, bottom = text_grob("Distance (km)"))
print(fs_ts)
}ggarrange(fluvsynchfun_mod(subbas = "West Brook", spear = synch_arr_sp_wb, ts = synch_arr_ts_wb,
dist_euc = dist_euclidean_wb, dist_wat = dist_watercourse_wb, dist_flow = dist_flowconn_wb),
fluvsynchfun_mod(subbas = "Staunton River", spear = synch_arr_sp_st, ts = synch_arr_ts_st,
dist_euc = dist_euclidean_st, dist_wat = dist_watercourse_st, dist_flow = dist_flowconn_st),
fluvsynchfun_mod(subbas = "Paine Run", spear = synch_arr_sp_pa, ts = synch_arr_ts_pa,
dist_euc = dist_euclidean_pa, dist_wat = dist_watercourse_pa, dist_flow = dist_flowconn_pa),
fluvsynchfun_mod(subbas = "Flathead River", spear = synch_arr_sp_fl, ts = synch_arr_ts_fl,
dist_euc = dist_euclidean_fl, dist_wat = dist_watercourse_fl, dist_flow = dist_flowconn_fl),
fluvsynchfun_mod(subbas = "Yellowstone River", spear = synch_arr_sp_ye, ts = synch_arr_ts_ye,
dist_euc = dist_euclidean_ye, dist_wat = dist_watercourse_ye, dist_flow = matrix(NA, nrow = nrow(dist_flowconn_ye), ncol = ncol(dist_flowconn_ye))),
fluvsynchfun_mod(subbas = "Snake River", spear = synch_arr_sp_sn, ts = synch_arr_ts_sn,
dist_euc = dist_euclidean_sn, dist_wat = dist_watercourse_sn, dist_flow = dist_flowconn_sn),
fluvsynchfun_mod(subbas = "Donner und Blitzen River", spear = synch_arr_sp_db, ts = synch_arr_ts_db,
dist_euc = dist_euclidean_db, dist_wat = dist_watercourse_db, dist_flow = dist_flowconn_db),
ncol = 1
)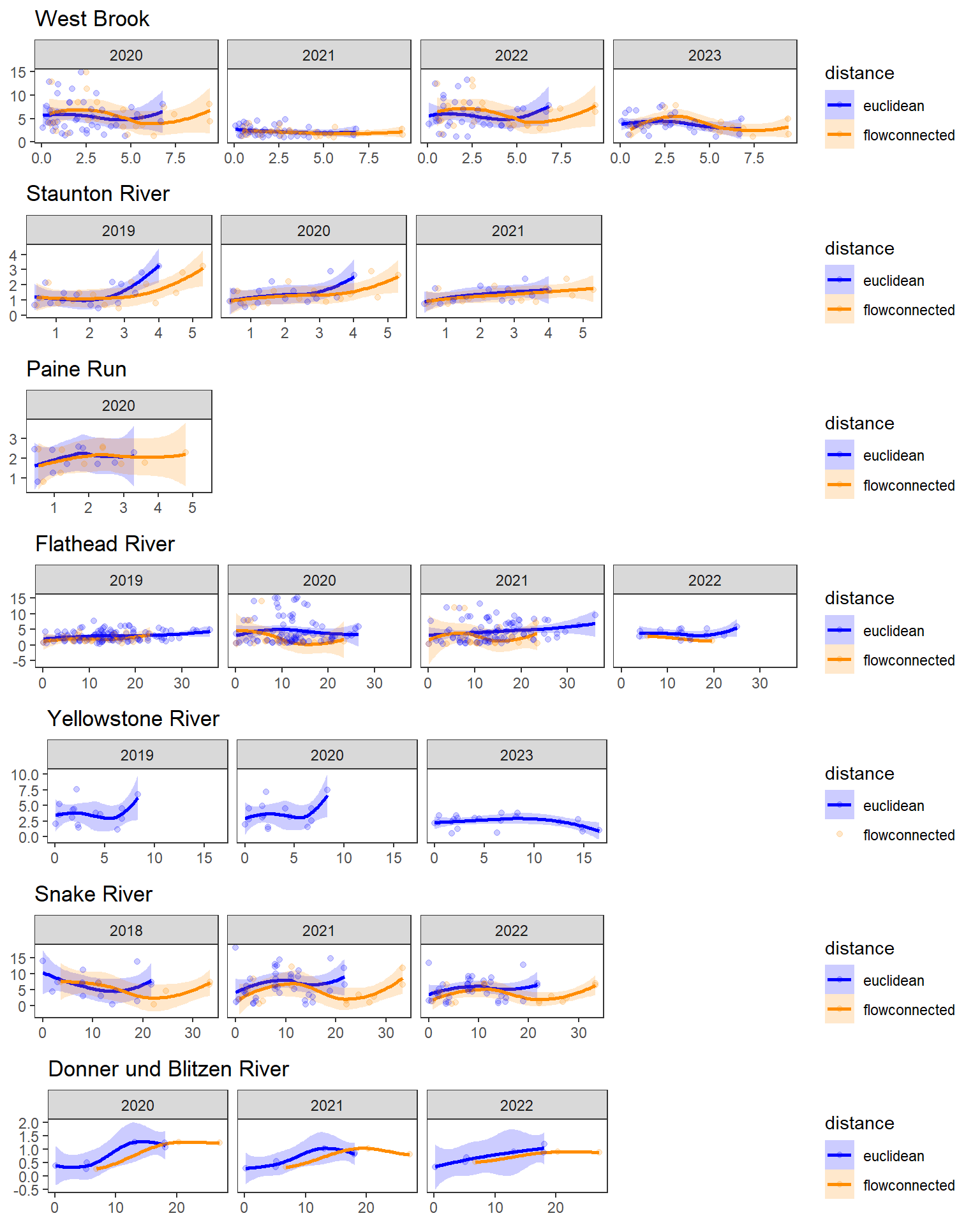
# jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/FluvialSynchrograms.jpg", width = 7, height = 12, units = "in", res = 1000)
# annotate_figure(p1, bottom = "Distance (km)", left = "Time series distance")
# dev.off()