Code
siteinfo <- read_csv("data/EcoDrought_SiteInformation.csv")
datatable(siteinfo)Code
siteinfo_sp <- st_as_sf(siteinfo, coords = c("long", "lat"), crs = 4326)
mapview(siteinfo_sp, zcol = "designation")Purpose: view EcoDrought stream temperature and flow data and download Daymet climate data (modeled air temperature).
siteinfo <- read_csv("data/EcoDrought_SiteInformation.csv")
datatable(siteinfo)siteinfo_sp <- st_as_sf(siteinfo, coords = c("long", "lat"), crs = 4326)
mapview(siteinfo_sp, zcol = "designation")Load data
dat <- read_csv("data/EcoDrought_FlowTempData_DailyWeekly.csv")
dat# A tibble: 202,014 × 31
station_no site_name site_id basin subbasin region lat long elev_ft
<chr> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
1 12355347 Big Creek NWIS BIG Flathe… Big Cre… Flat 48.6 -114. 3528.
2 12355347 Big Creek NWIS BIG Flathe… Big Cre… Flat 48.6 -114. 3528.
3 12355347 Big Creek NWIS BIG Flathe… Big Cre… Flat 48.6 -114. 3528.
4 12355347 Big Creek NWIS BIG Flathe… Big Cre… Flat 48.6 -114. 3528.
5 12355347 Big Creek NWIS BIG Flathe… Big Cre… Flat 48.6 -114. 3528.
6 12355347 Big Creek NWIS BIG Flathe… Big Cre… Flat 48.6 -114. 3528.
7 12355347 Big Creek NWIS BIG Flathe… Big Cre… Flat 48.6 -114. 3528.
8 12355347 Big Creek NWIS BIG Flathe… Big Cre… Flat 48.6 -114. 3528.
9 12355347 Big Creek NWIS BIG Flathe… Big Cre… Flat 48.6 -114. 3528.
10 12355347 Big Creek NWIS BIG Flathe… Big Cre… Flat 48.6 -114. 3528.
# ℹ 202,004 more rows
# ℹ 22 more variables: area_sqmi <dbl>, designation <chr>, date <date>,
# DischargeReliability <dbl>, TempReliability <dbl>, flow_mean <dbl>,
# flow_min <dbl>, flow_max <dbl>, tempc_mean <dbl>, tempc_min <dbl>,
# tempc_max <dbl>, flow_mean_filled <dbl>, flow_mean_cms <dbl>,
# flow_mean_filled_cms <dbl>, area_sqkm <dbl>, Yield_mm <dbl>,
# Yield_filled_mm <dbl>, flow_mean_7 <dbl>, flow_mean_filled_7 <dbl>, …Unique basins
unique(dat$basin)[1] "Flathead" "West Brook" "Donner Blitzen" "Paine Run"
[5] "Piney River" "Staunton River" "Shields River" "Snake River" Trim siteinfo to sites with temp/flow data
siteinfo_sub <- siteinfo %>% filter(site_name %in% unique(dat$site_name))Download Daymet daily air temperature data for each site (point locations)
climlist <- vector("list", length = dim(siteinfo_sub)[1])
for (i in 1:dim(siteinfo_sub)[1]) {
clim <- download_daymet(site = siteinfo_sub$site_name[i], lat = siteinfo_sub$lat[i], lon = siteinfo_sub$long[i], start = 2010, end = 2024, internal = T)
climlist[[i]] <- tibble(clim$data) %>%
mutate(air_temp_mean = (tmax..deg.c. + tmin..deg.c.)/2,
date = as.Date(paste(year, yday, sep = "-"), "%Y-%j"),
site_name = siteinfo_sub$site_name[i]) %>%
select(12,2,11,10,4,6,3,5) %>% rename(precip_mmday = 5, swe_kgm2 = 6, daylength_sec = 7, shortrad_wm2 = 8)
print(i)
}
climdf <- do.call(rbind, climlist)
write_csv(climdf, "data/Daymet_daily.csv")Re-load Daymet climate data
climdf <- read_csv("data/Daymet_daily.csv")Plot example Daymet time series data (2020-2025, Avery Brook, MA)
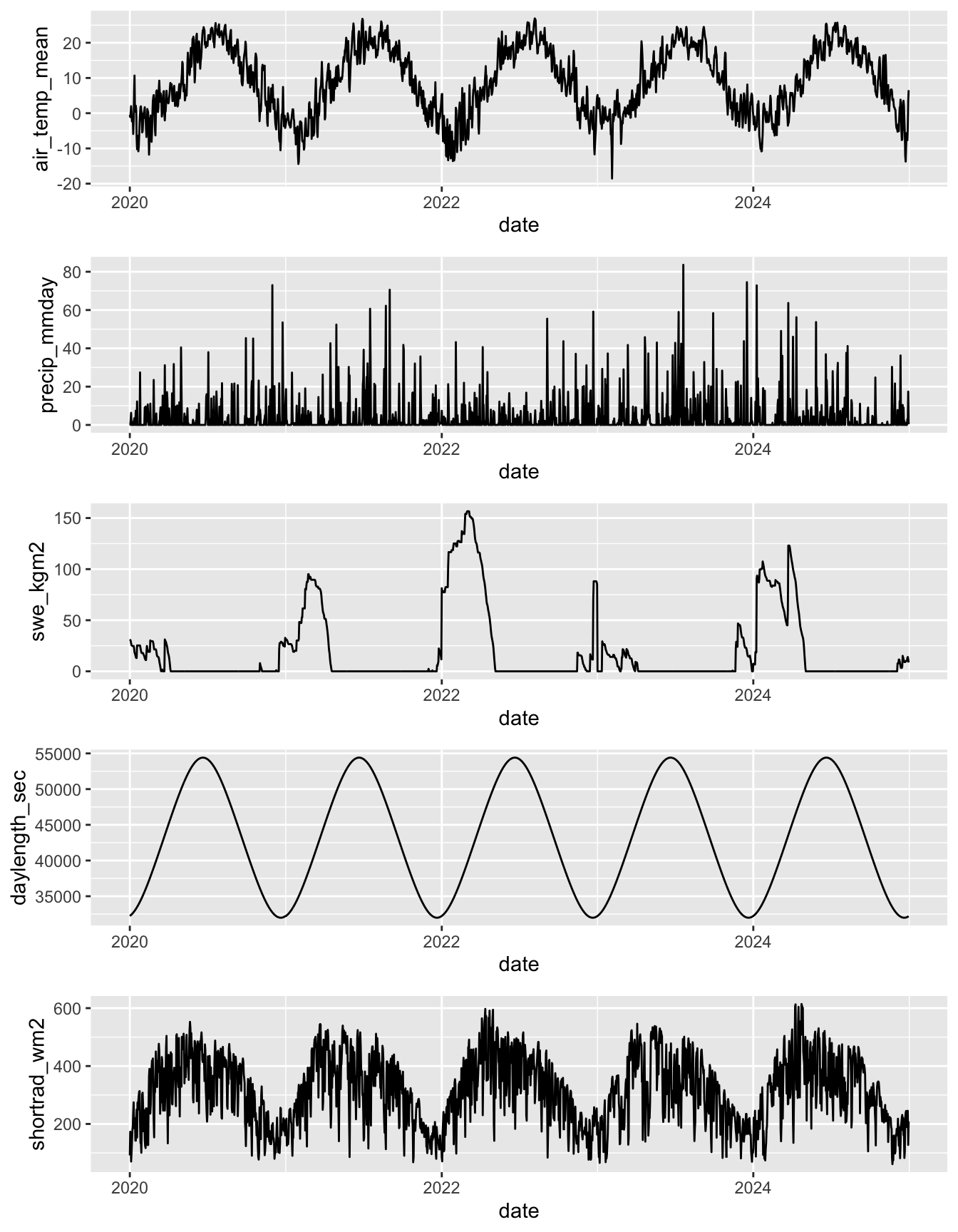
ggarrange(climdf %>% filter(site_name == "Avery Brook", year(date) >= 2020) %>% ggplot(aes(x = date, y = air_temp_mean)) + geom_line(),
climdf %>% filter(site_name == "Avery Brook", year(date) >= 2020) %>% ggplot(aes(x = date, y = precip_mmday)) + geom_line(),
climdf %>% filter(site_name == "Avery Brook", year(date) >= 2020) %>% ggplot(aes(x = date, y = swe_kgm2)) + geom_line(),
climdf %>% filter(site_name == "Avery Brook", year(date) >= 2020) %>% ggplot(aes(x = date, y = daylength_sec)) + geom_line(),
climdf %>% filter(site_name == "Avery Brook", year(date) >= 2020) %>% ggplot(aes(x = date, y = shortrad_wm2)) + geom_line(),
ncol = 1)