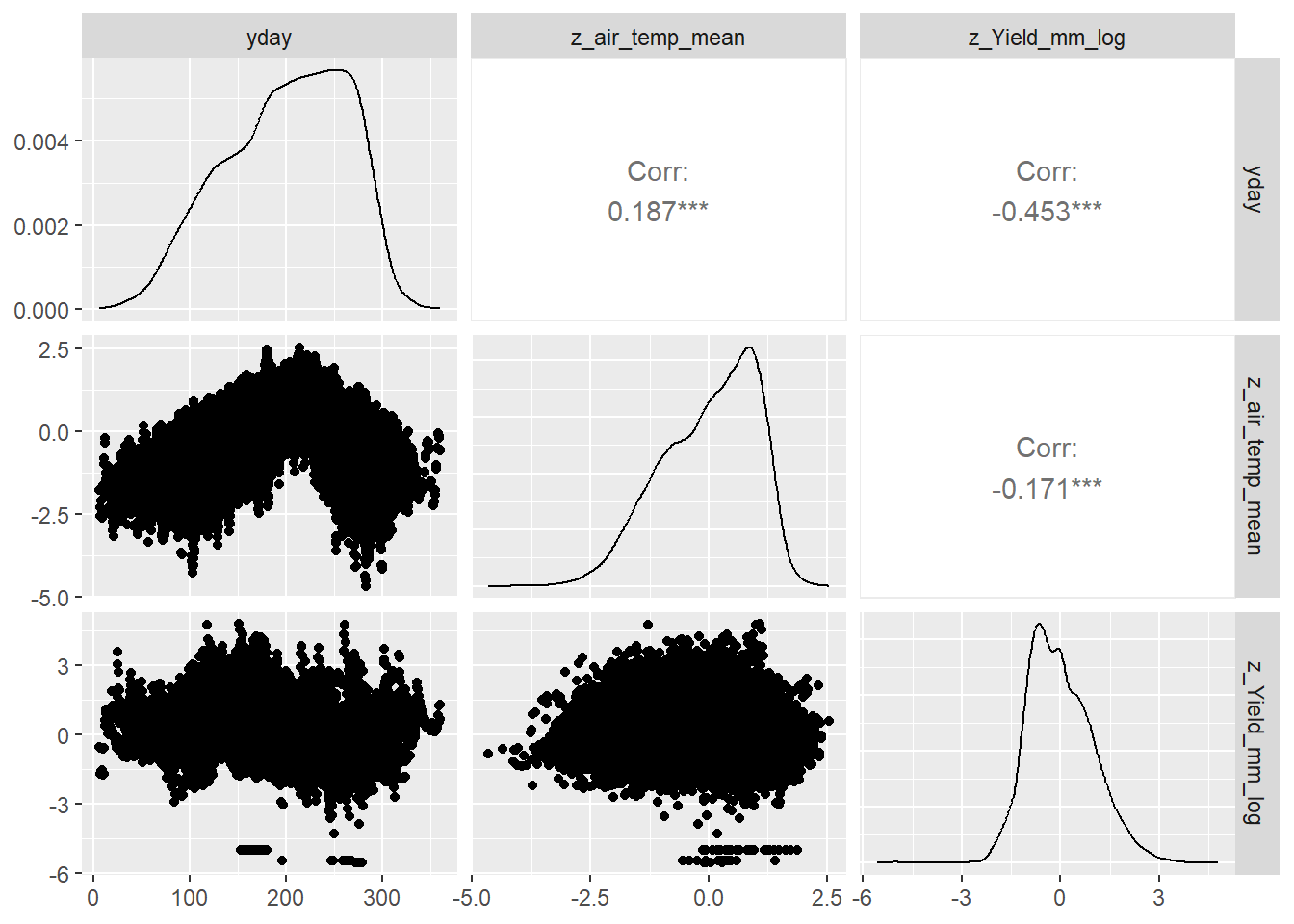
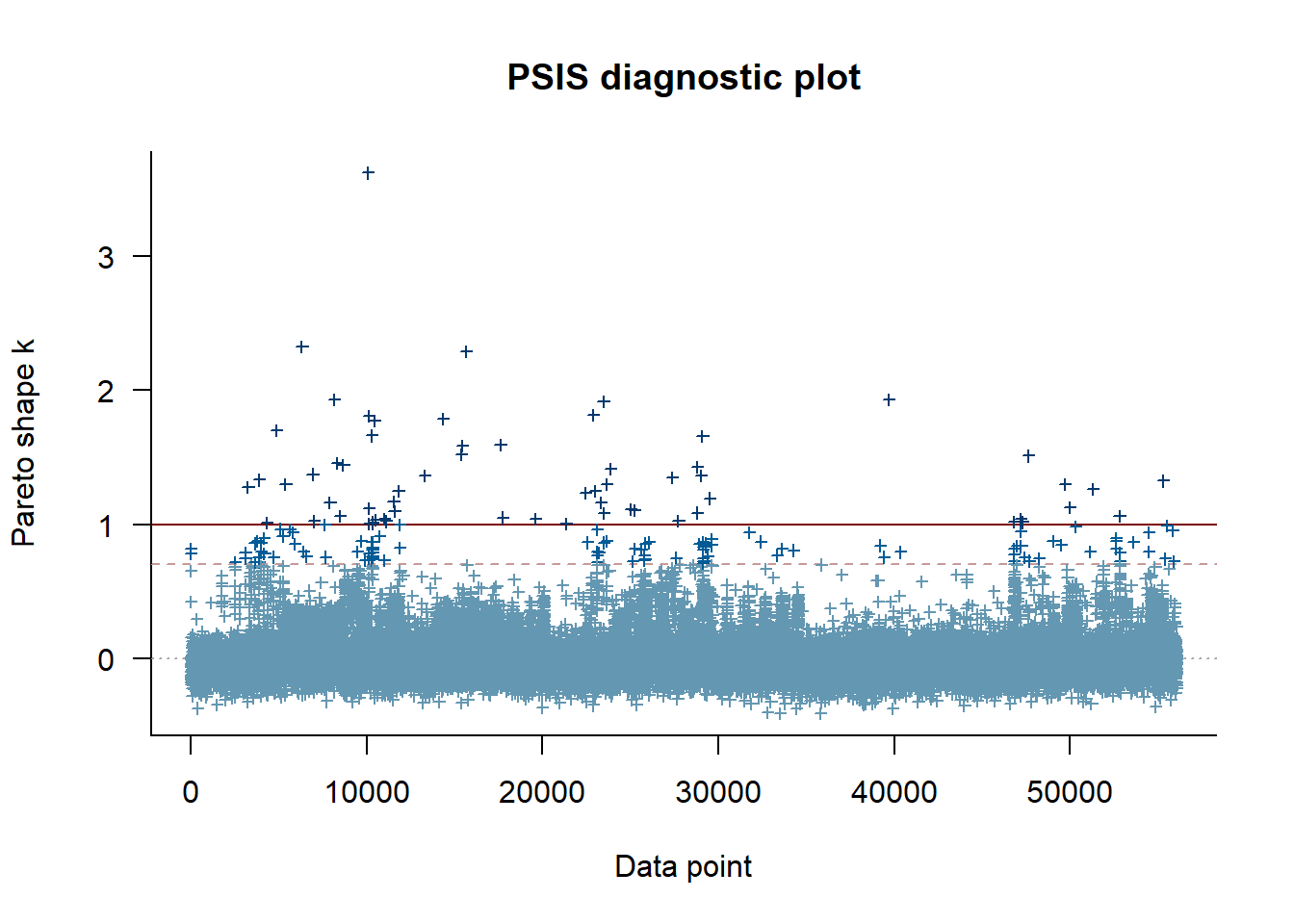
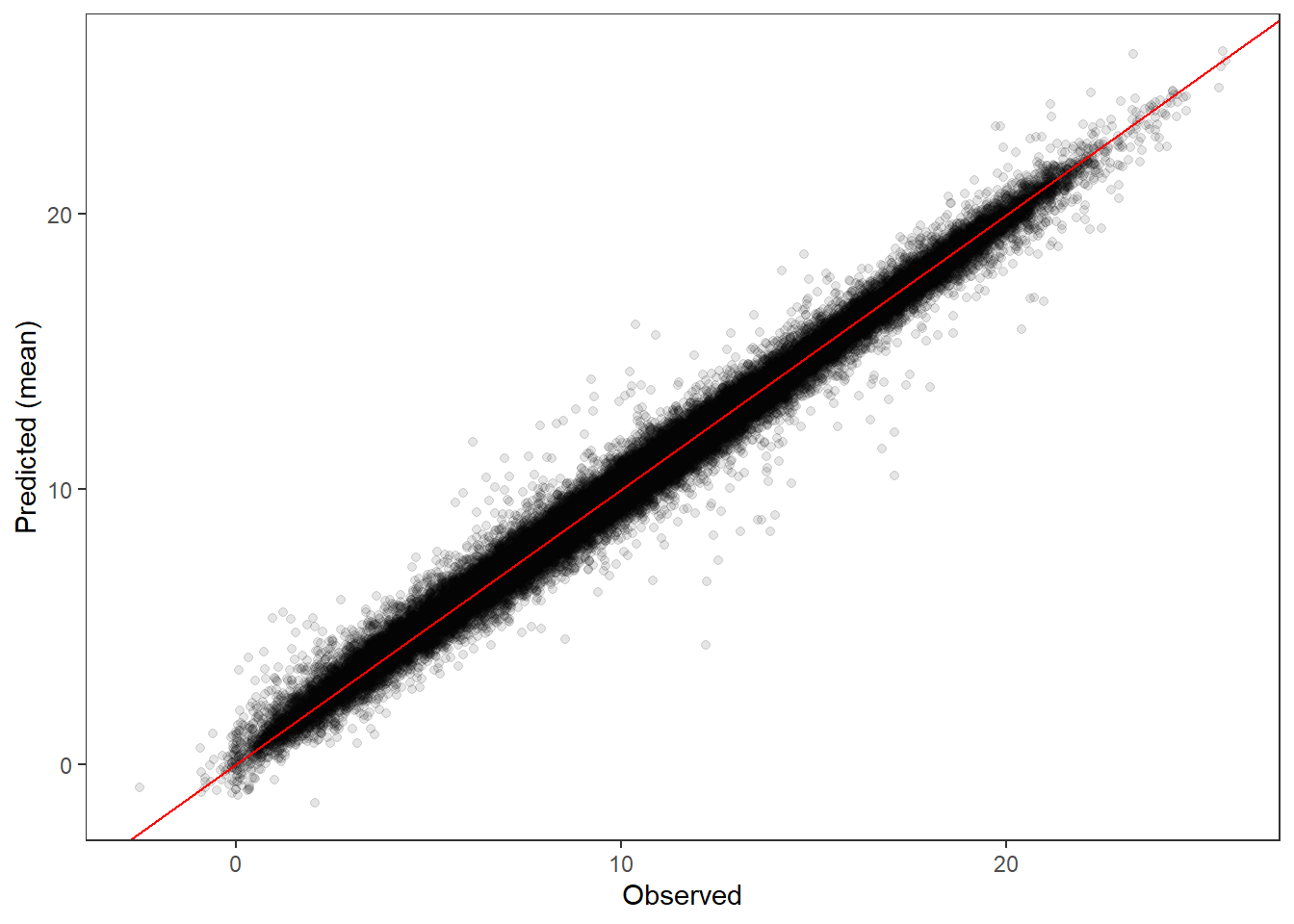
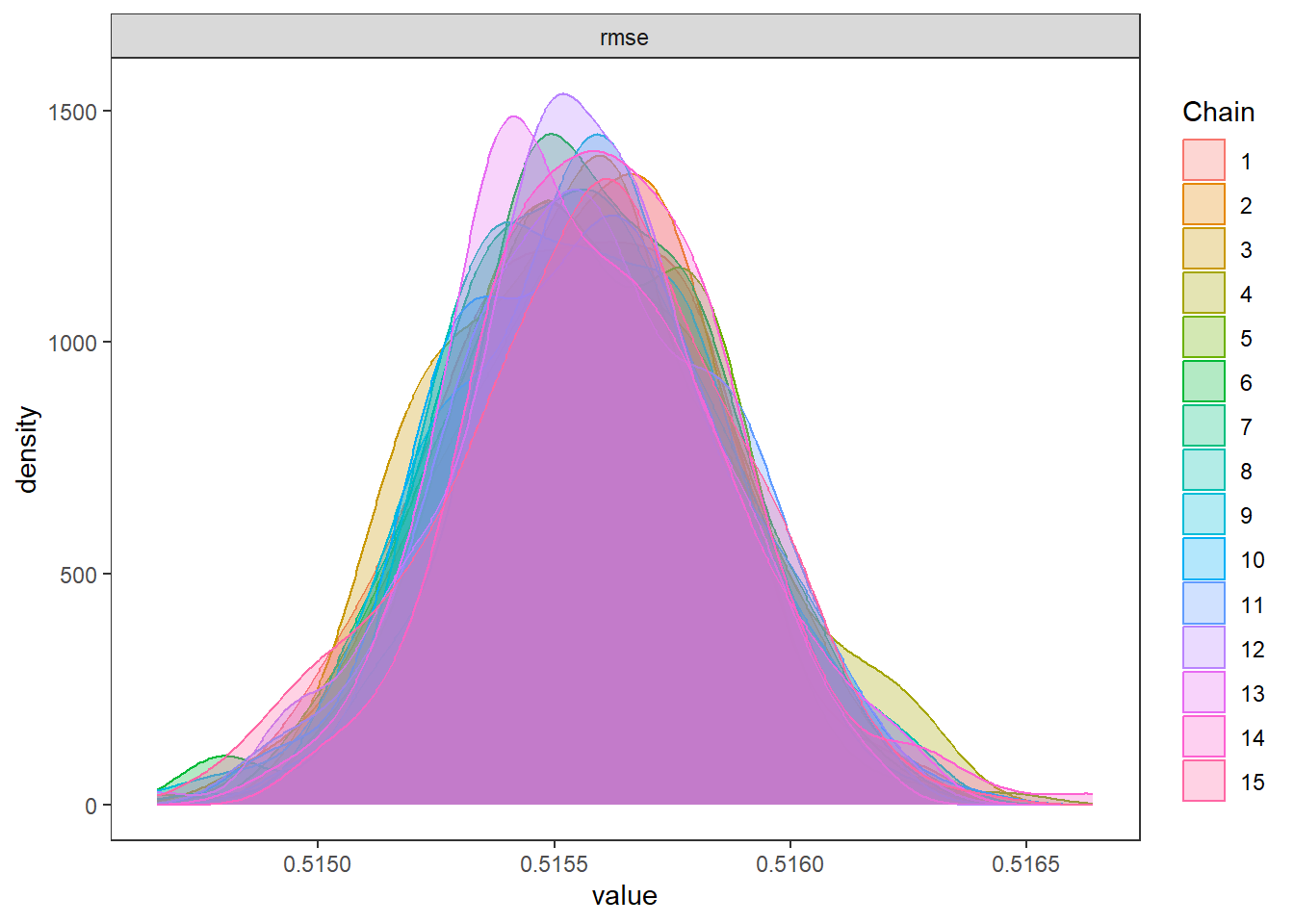
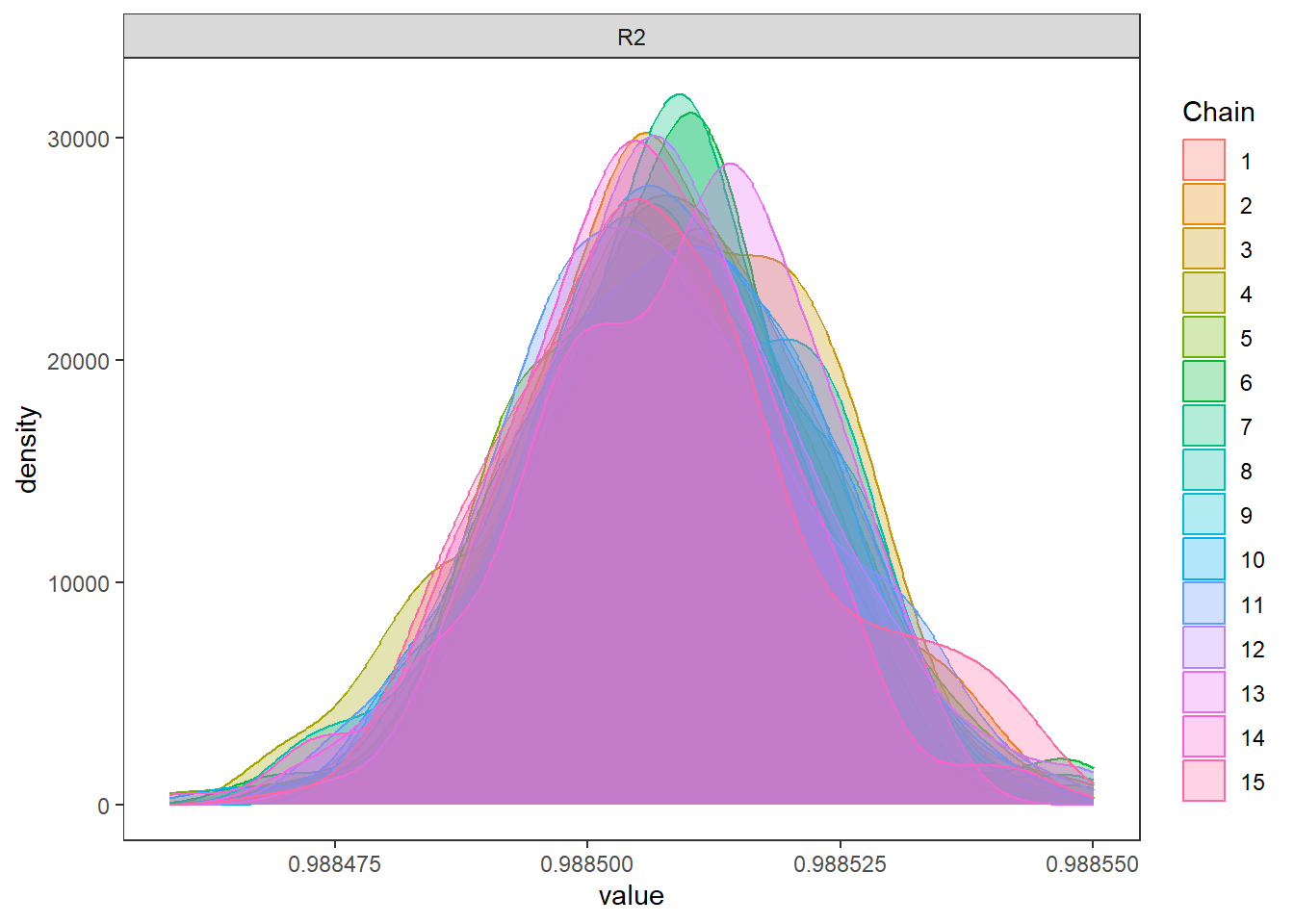
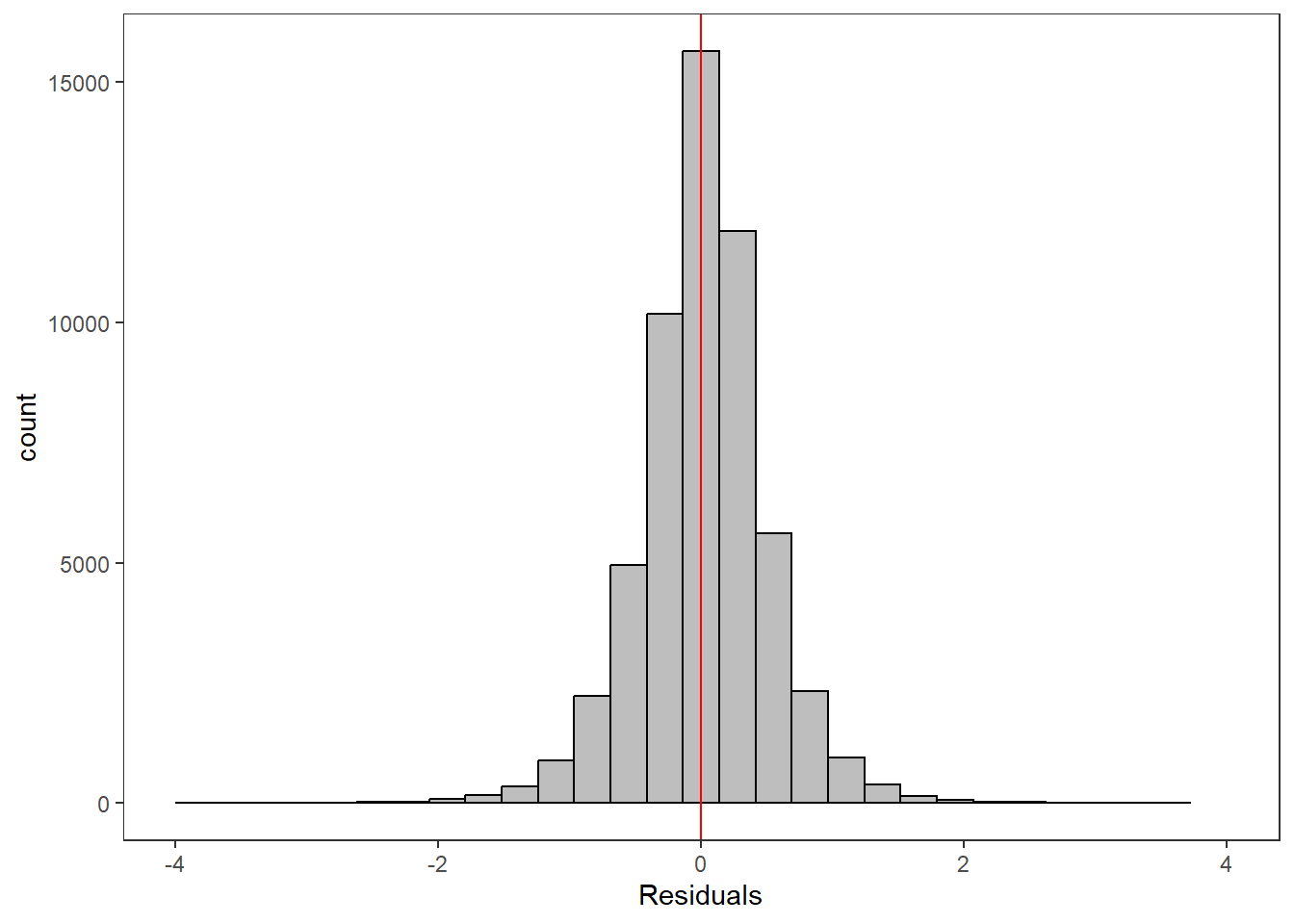
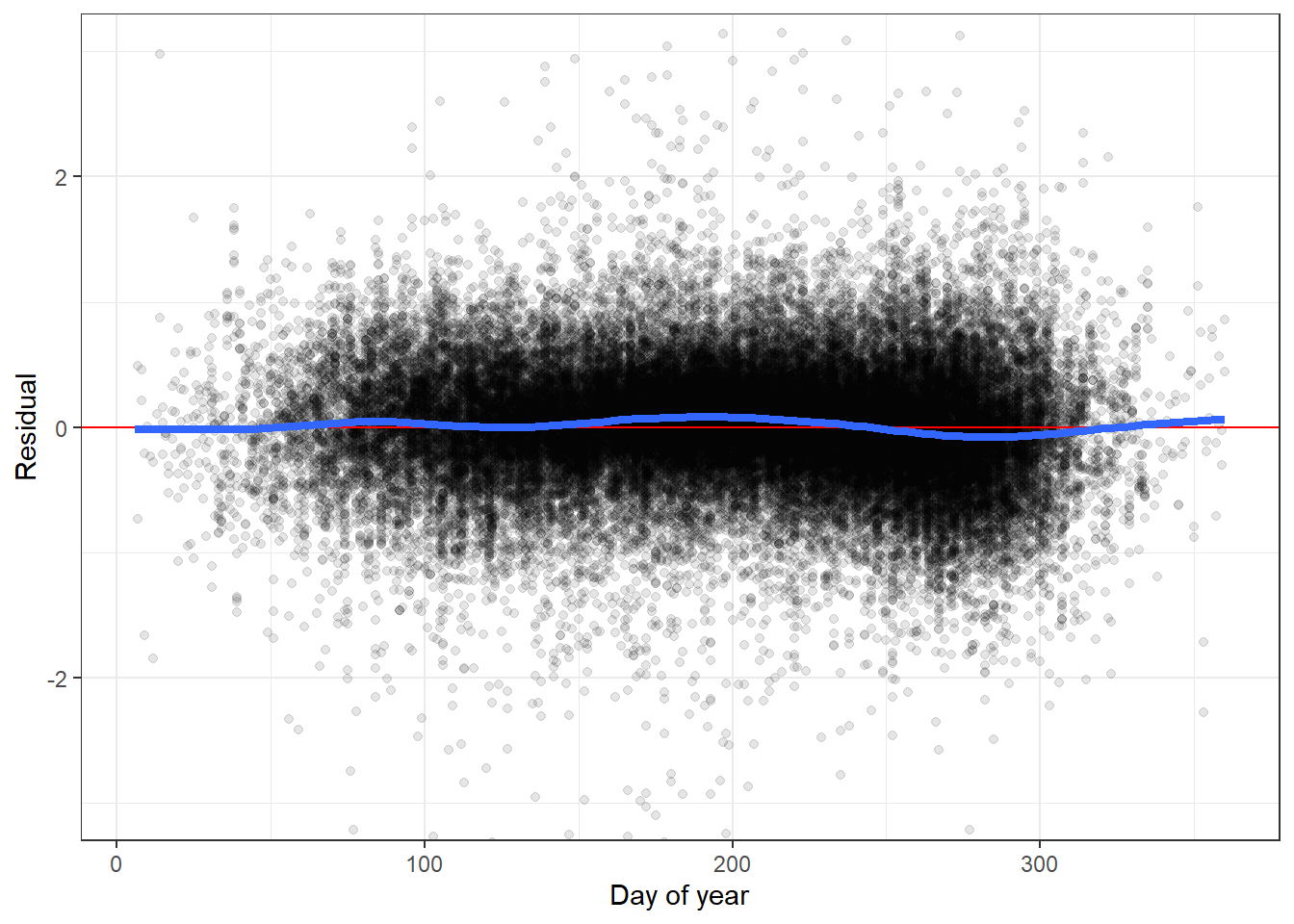
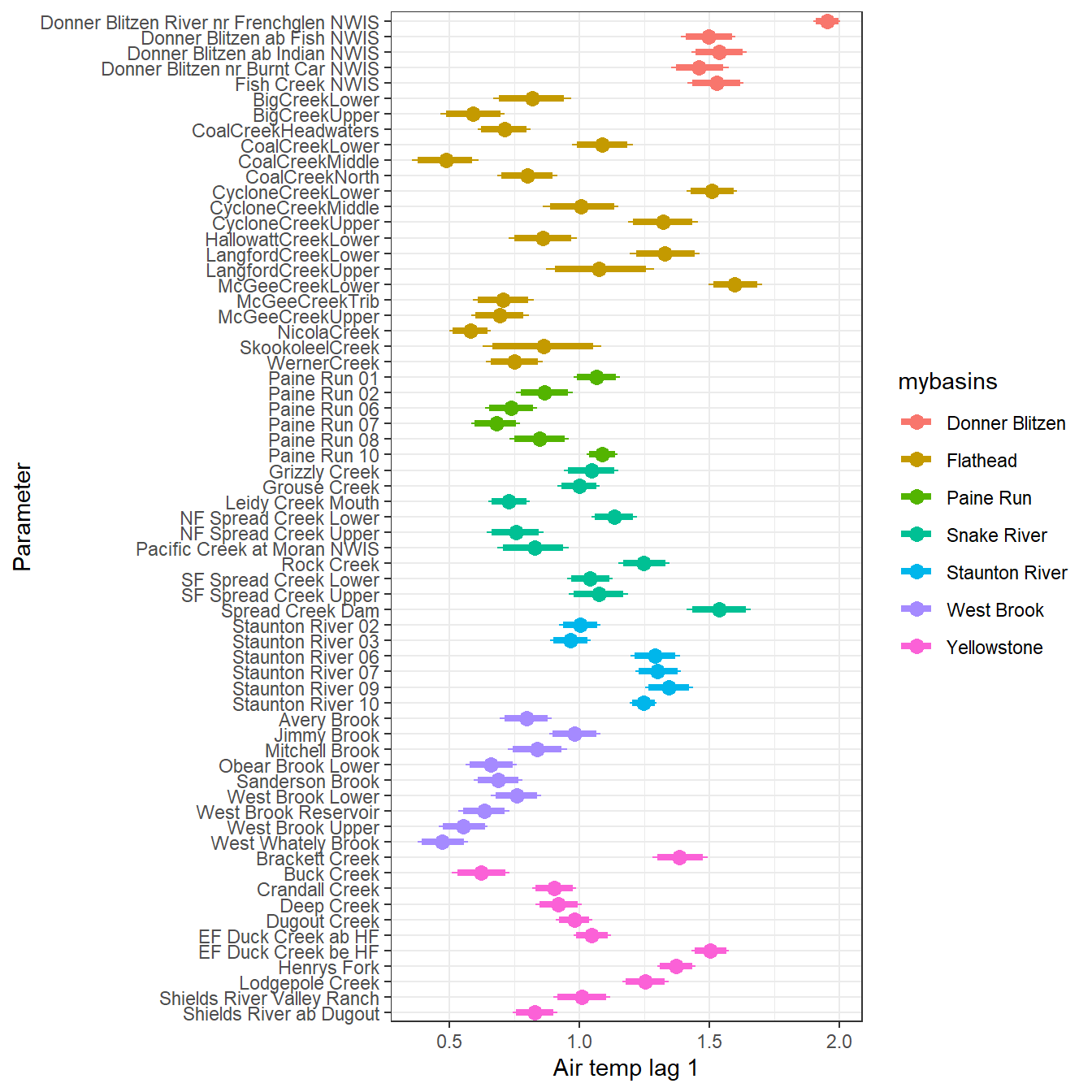
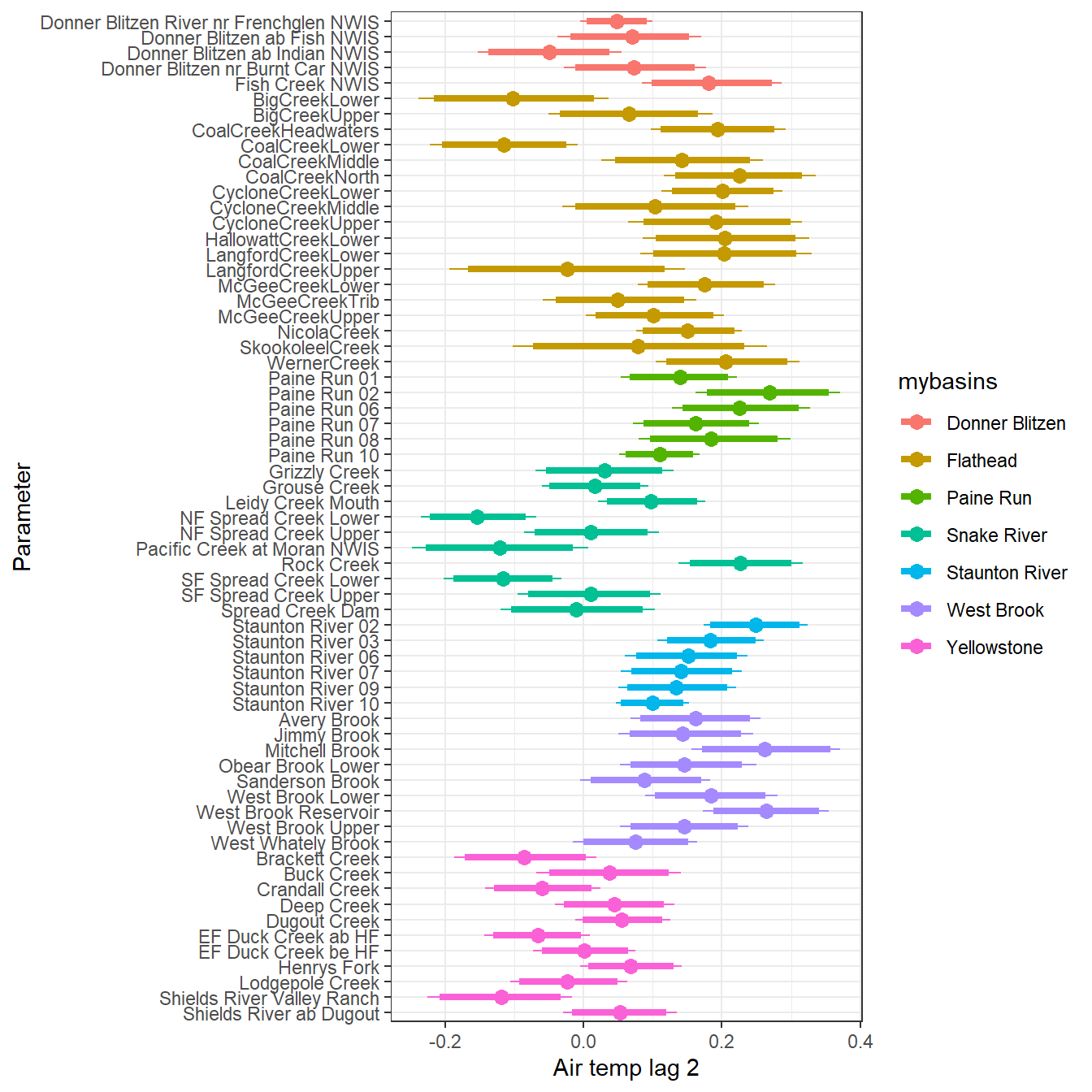
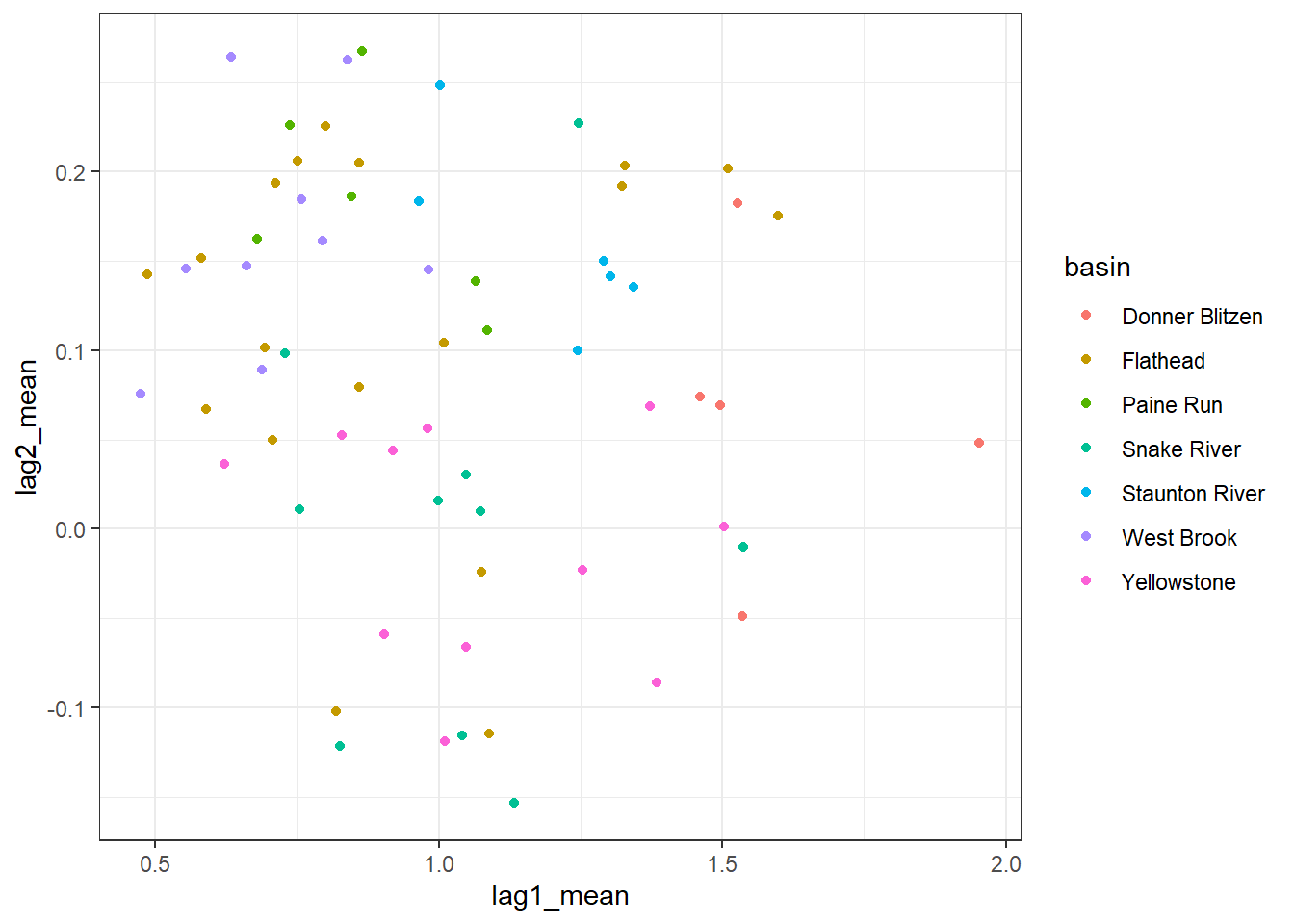
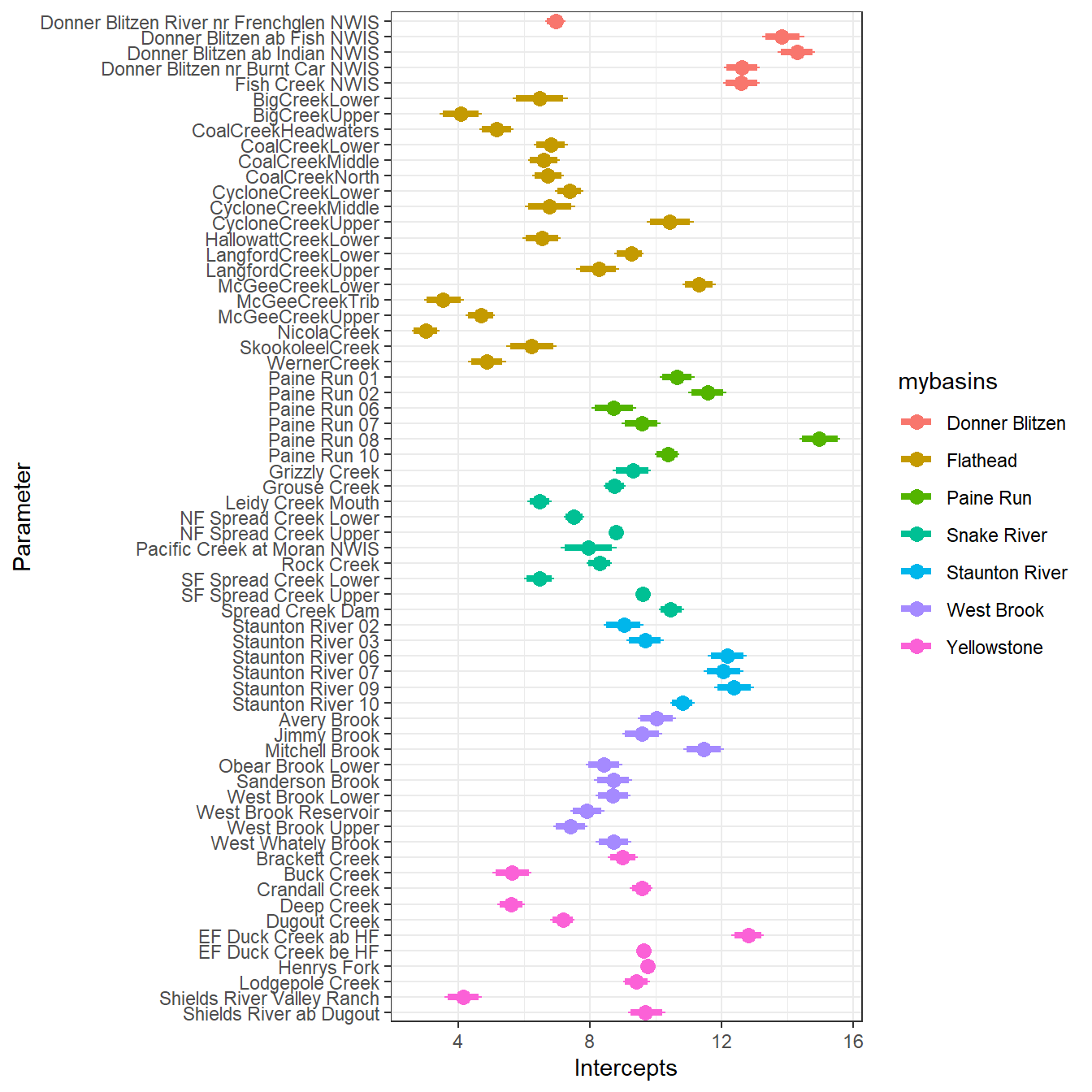
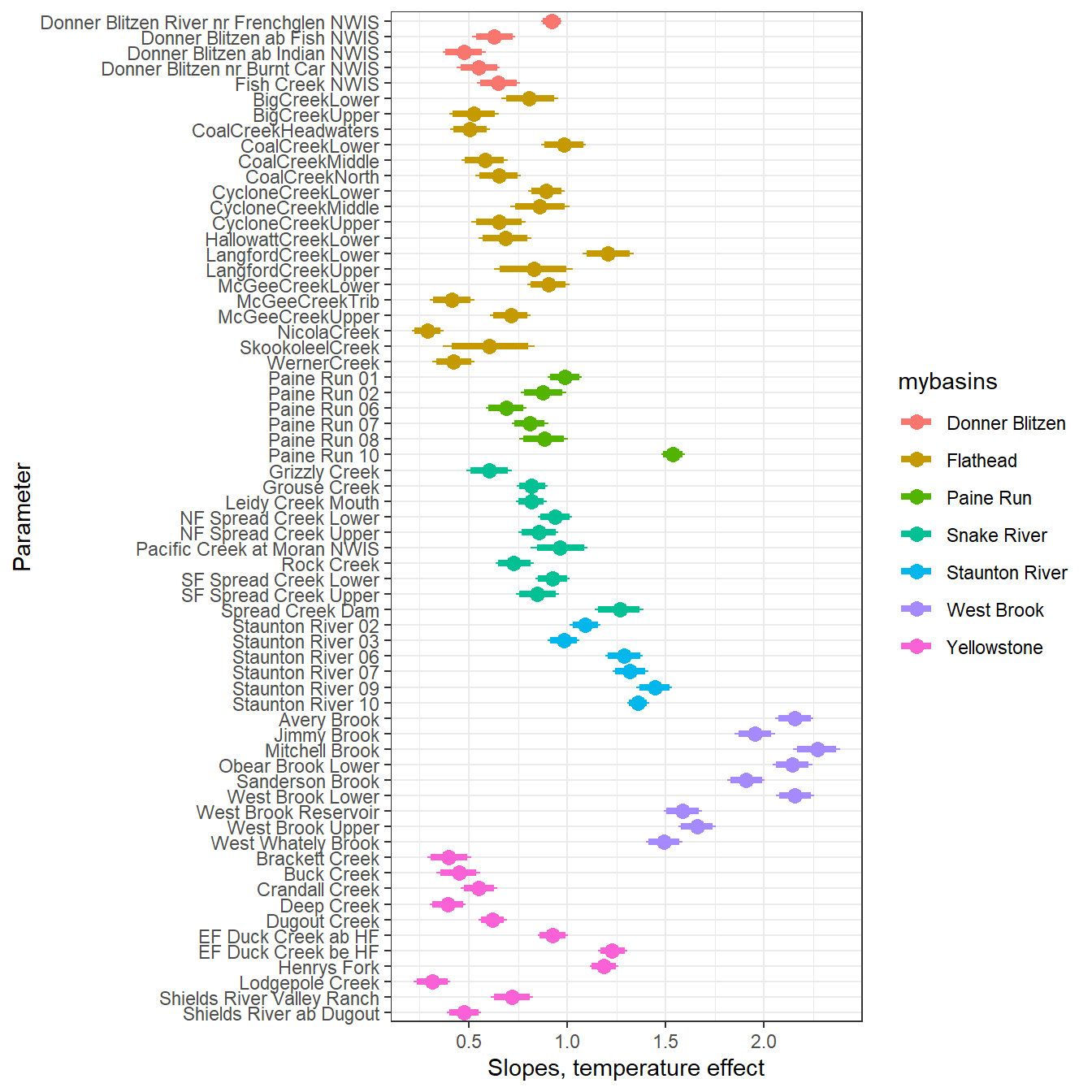
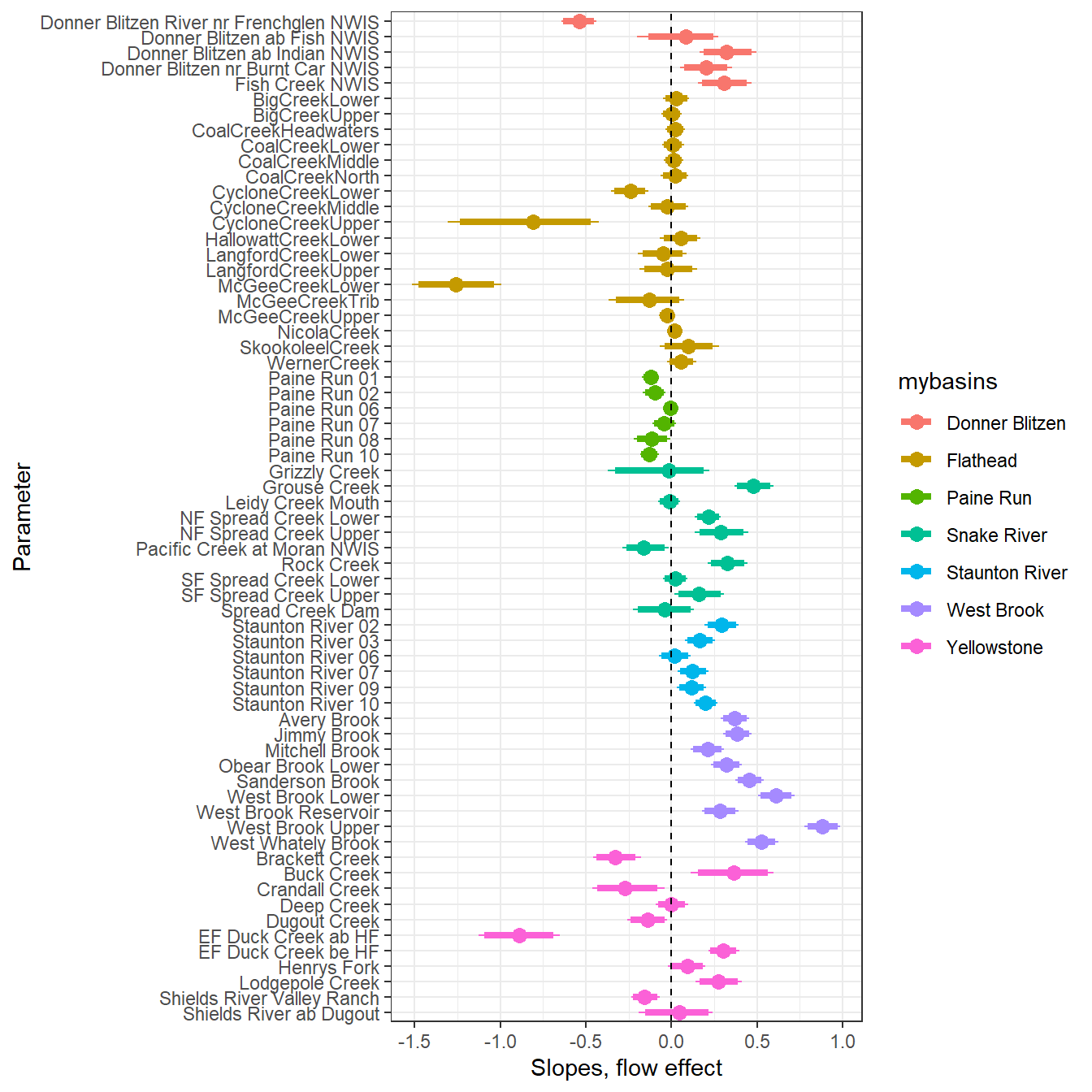
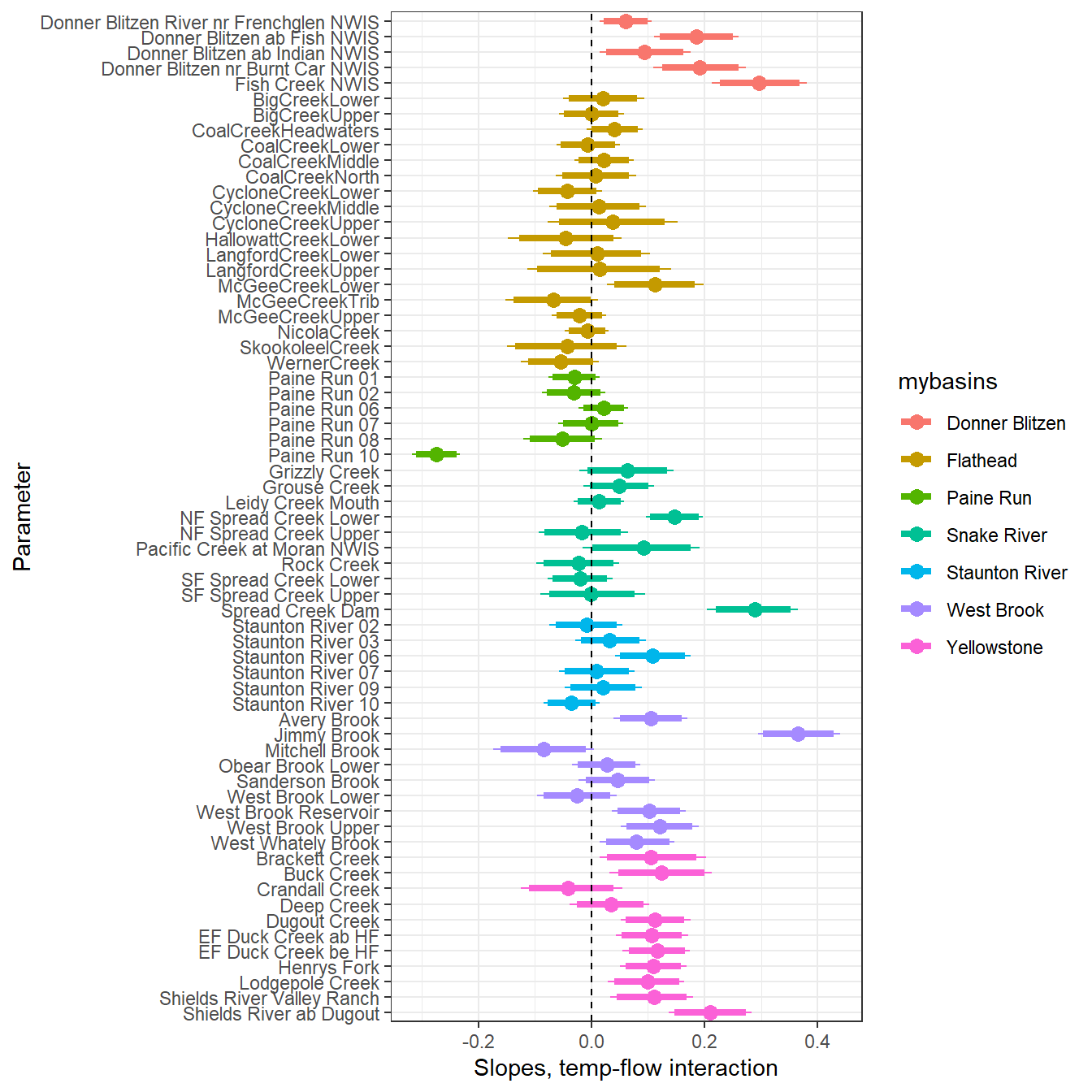
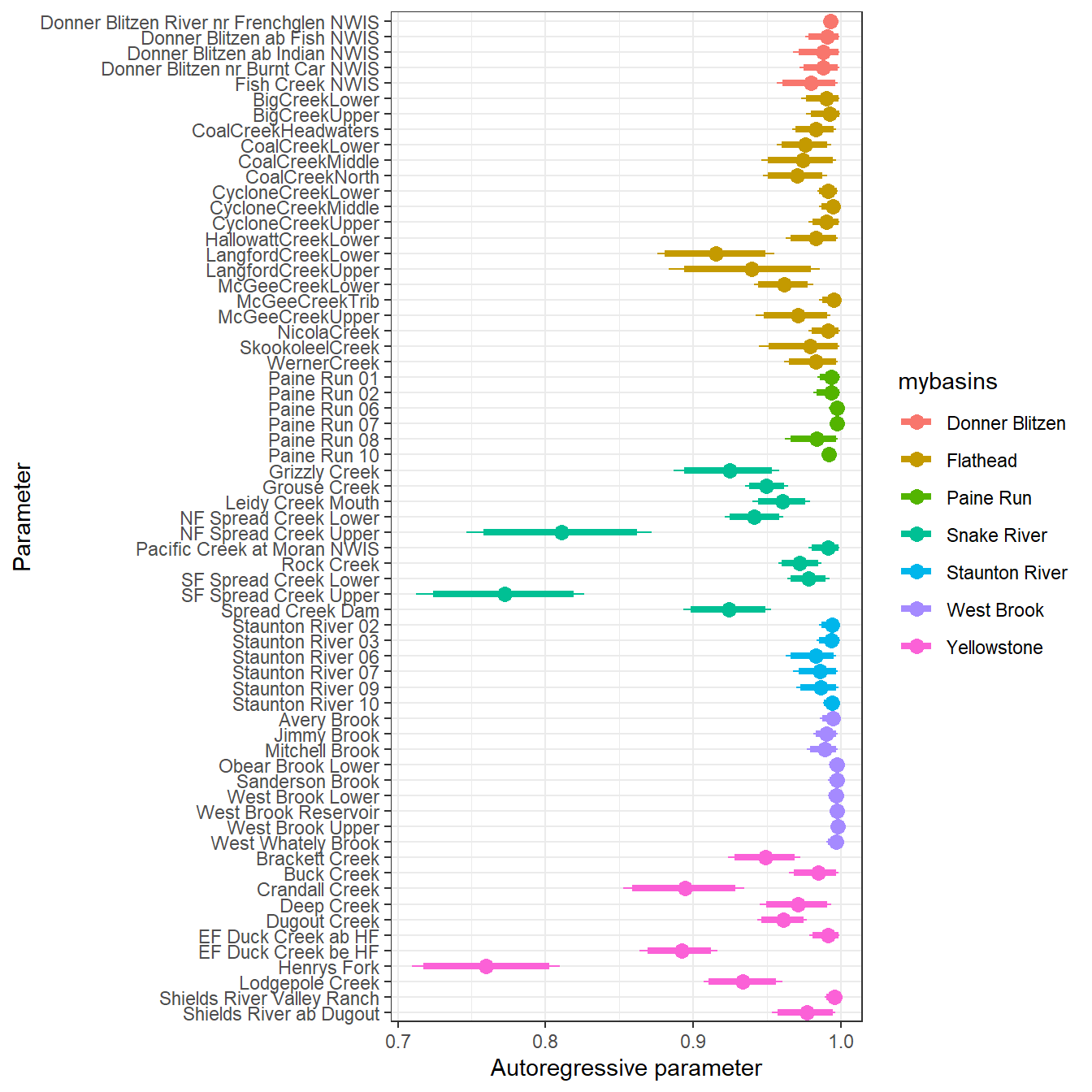
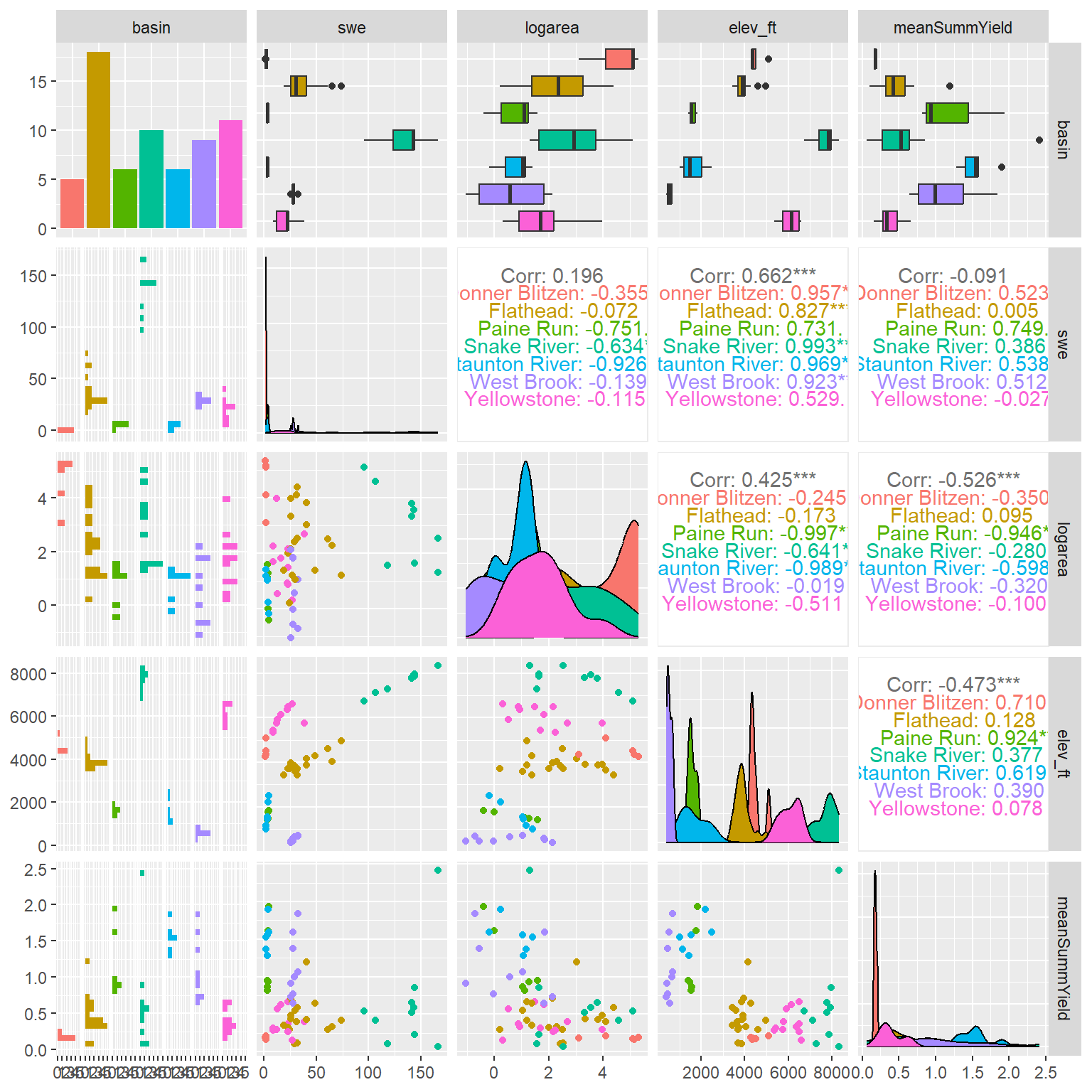
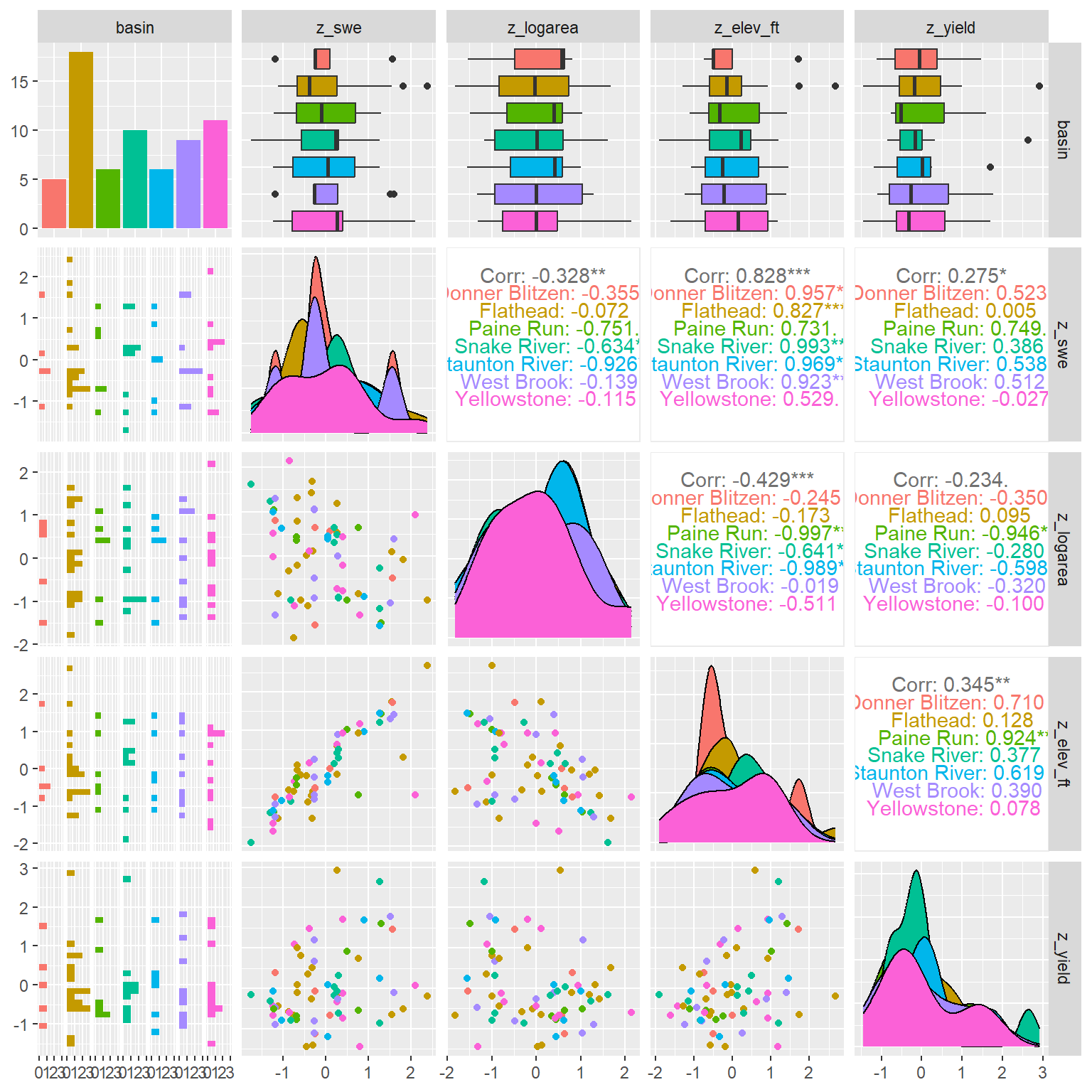
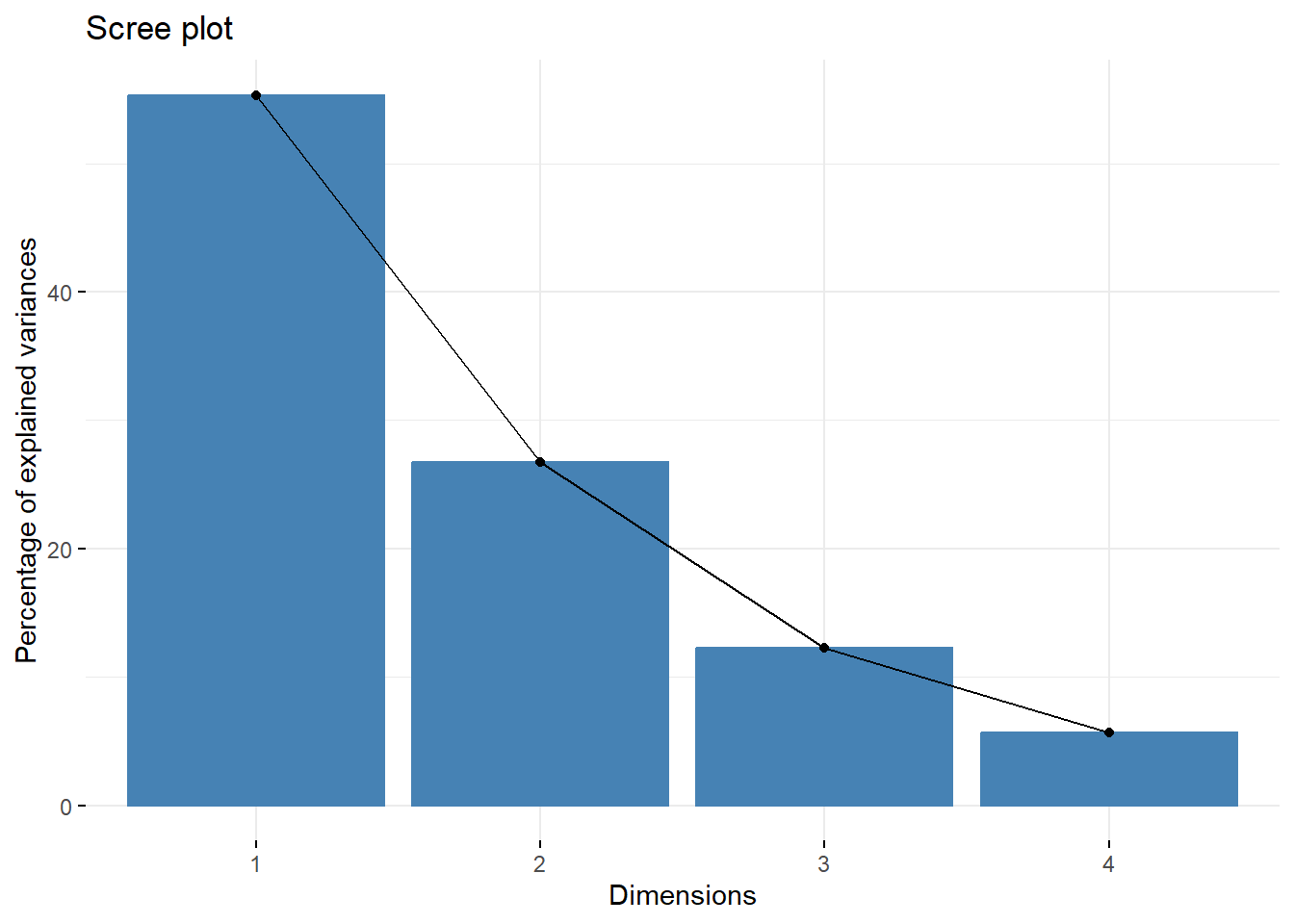
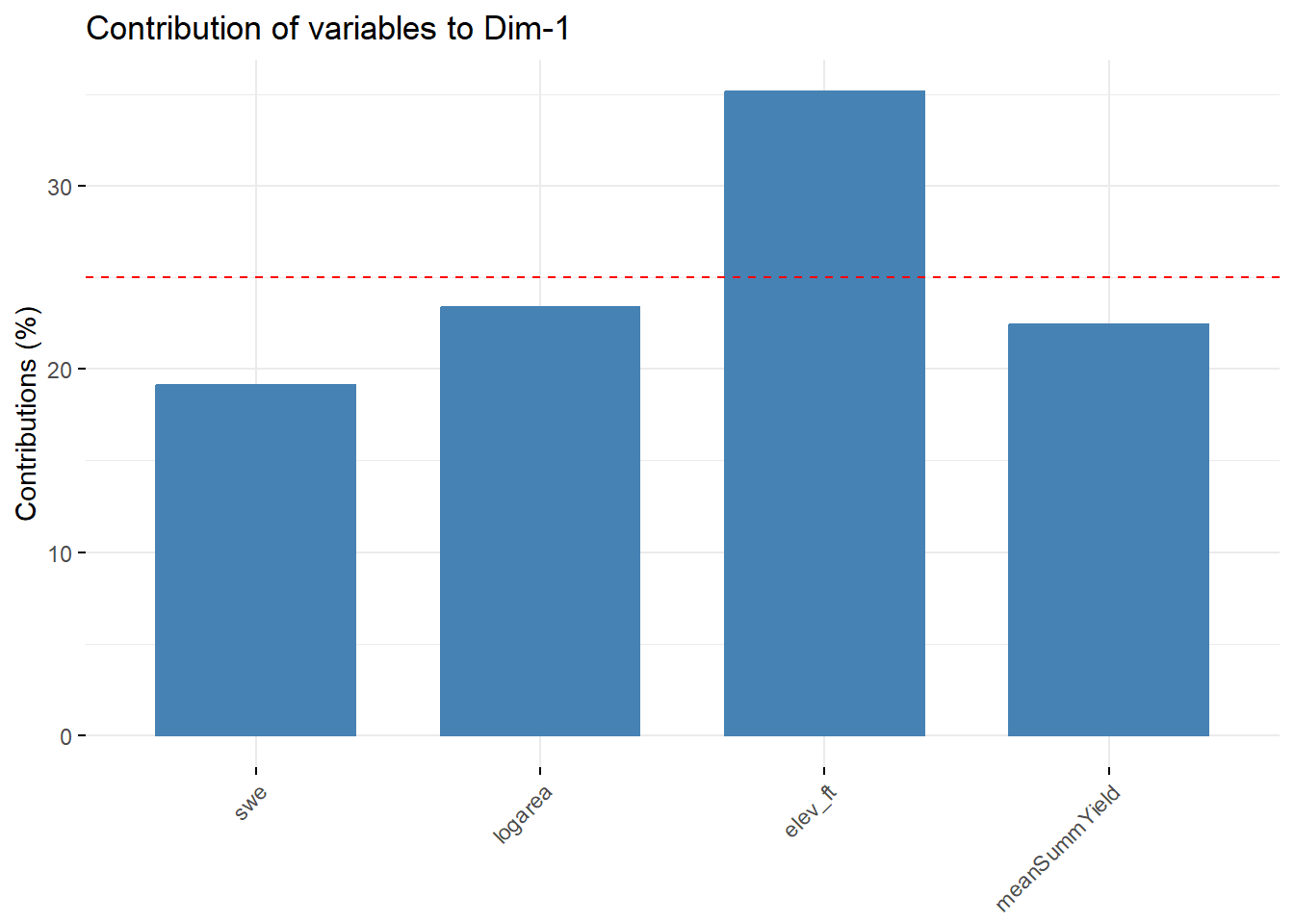
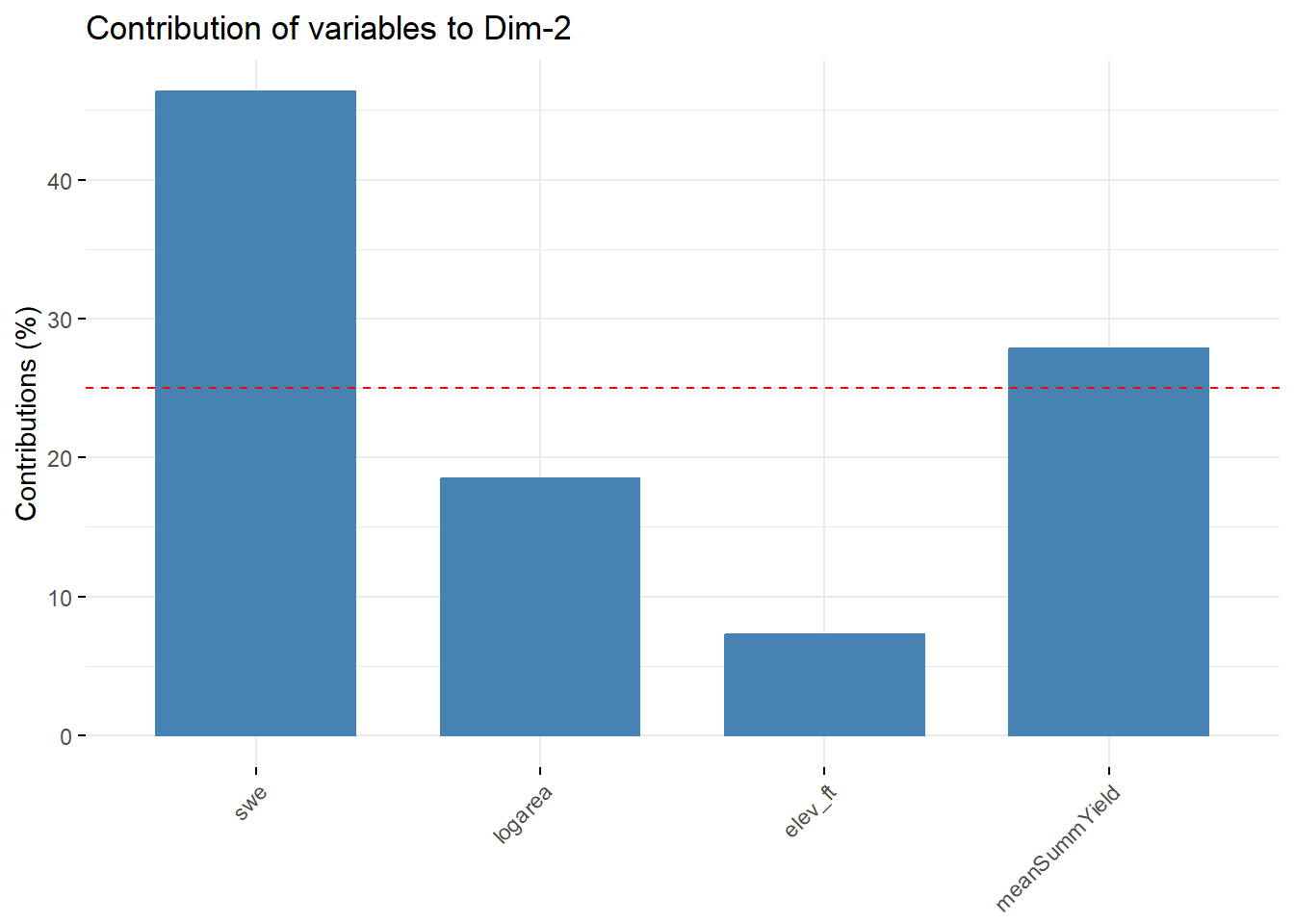
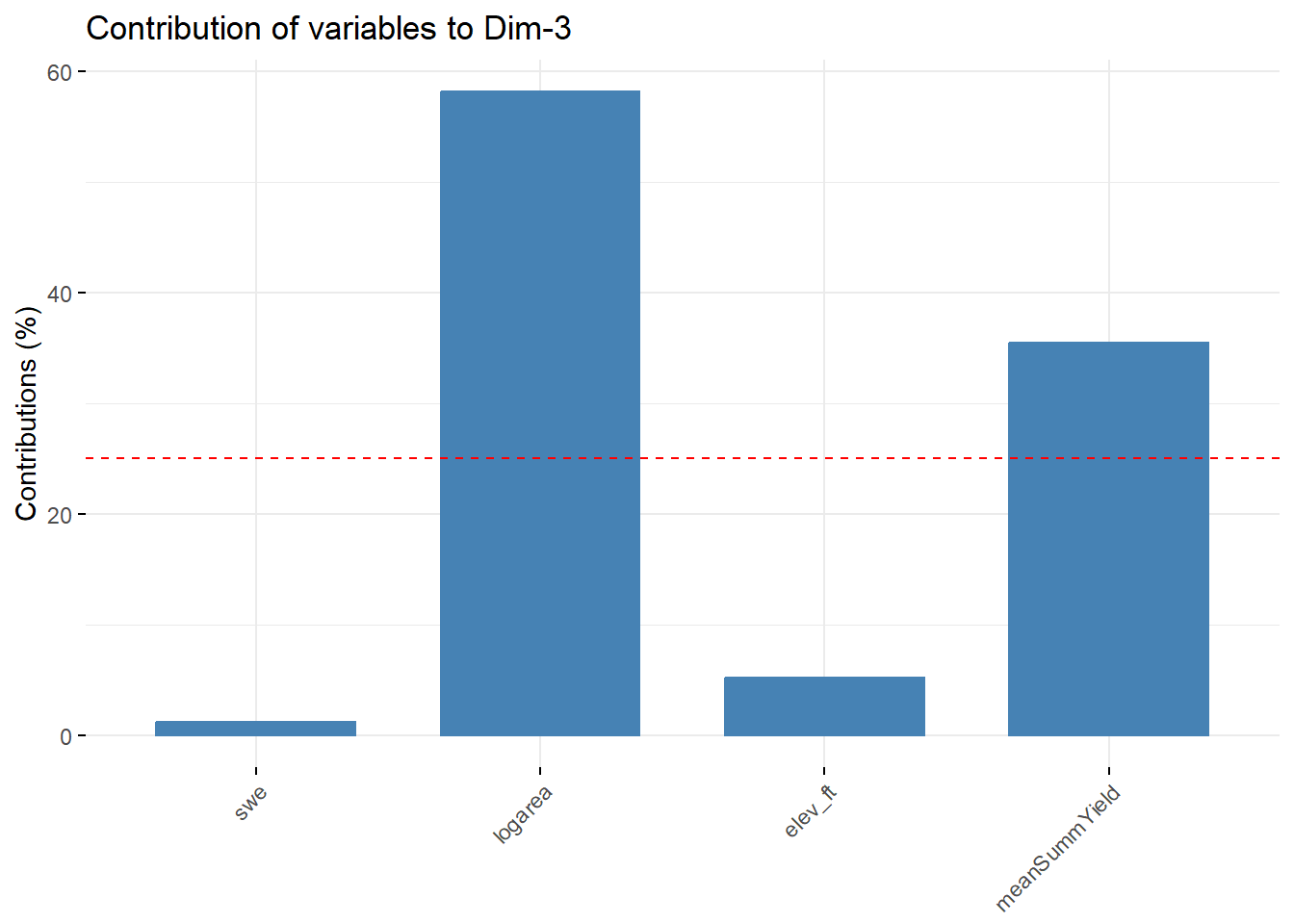
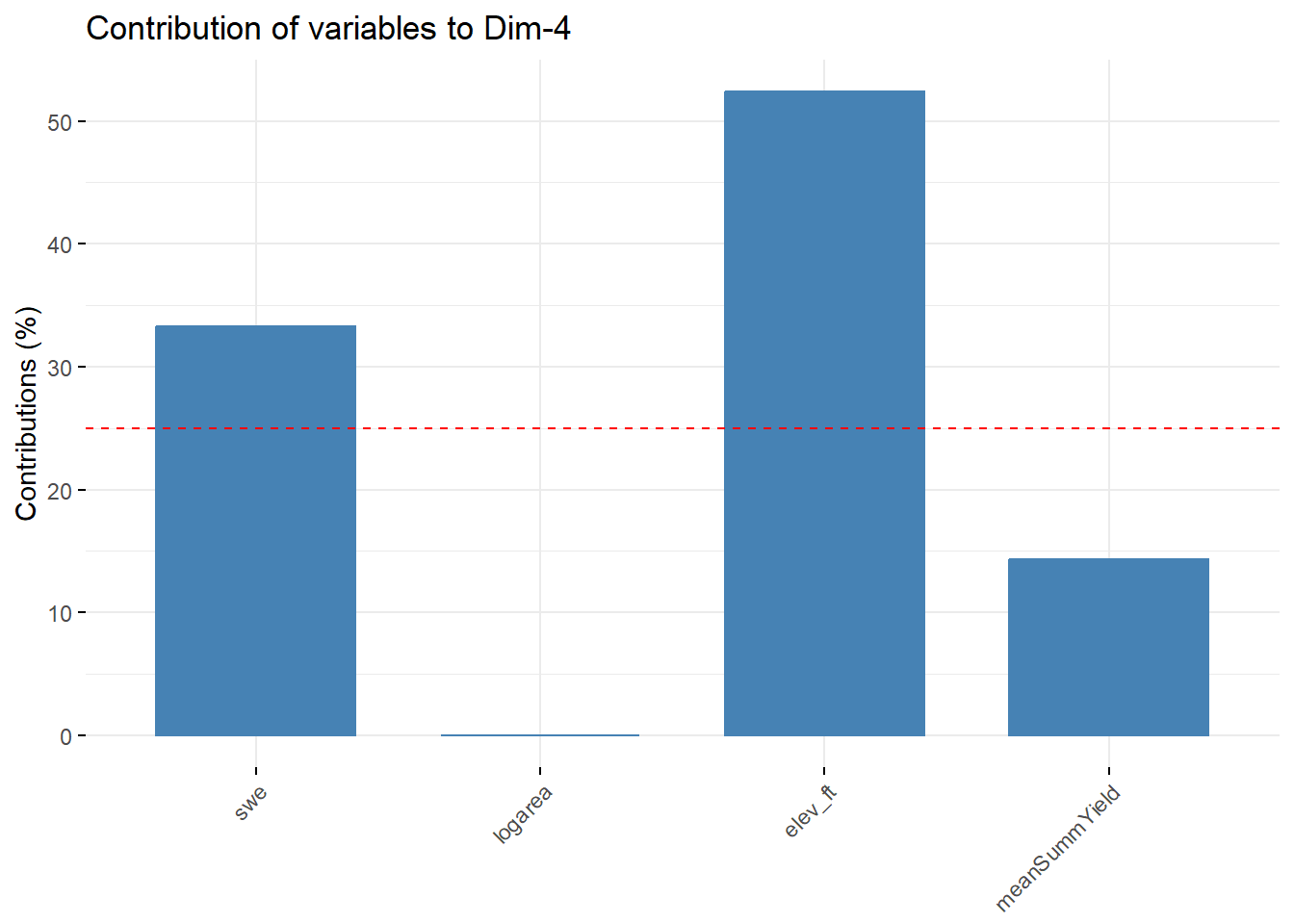
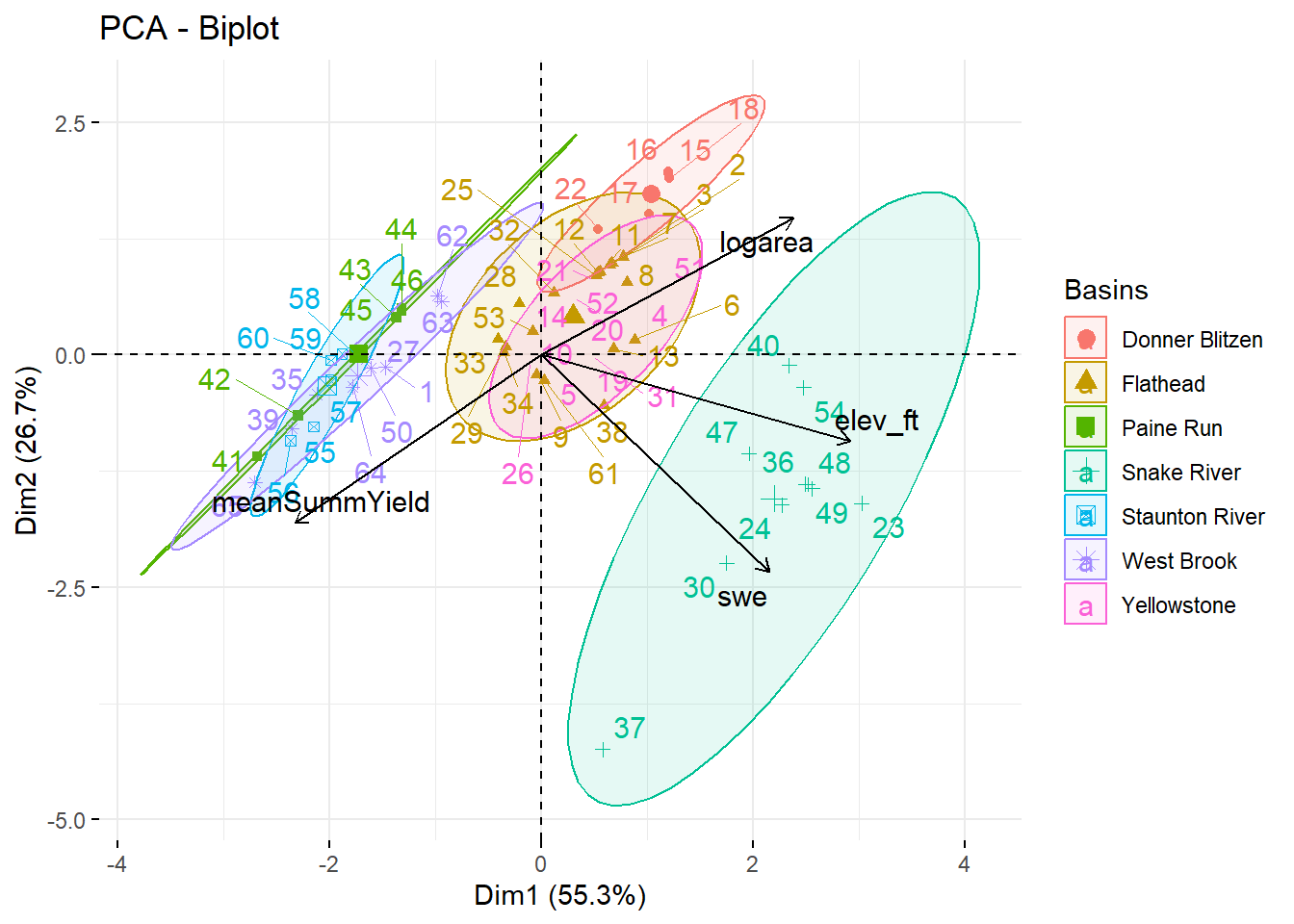
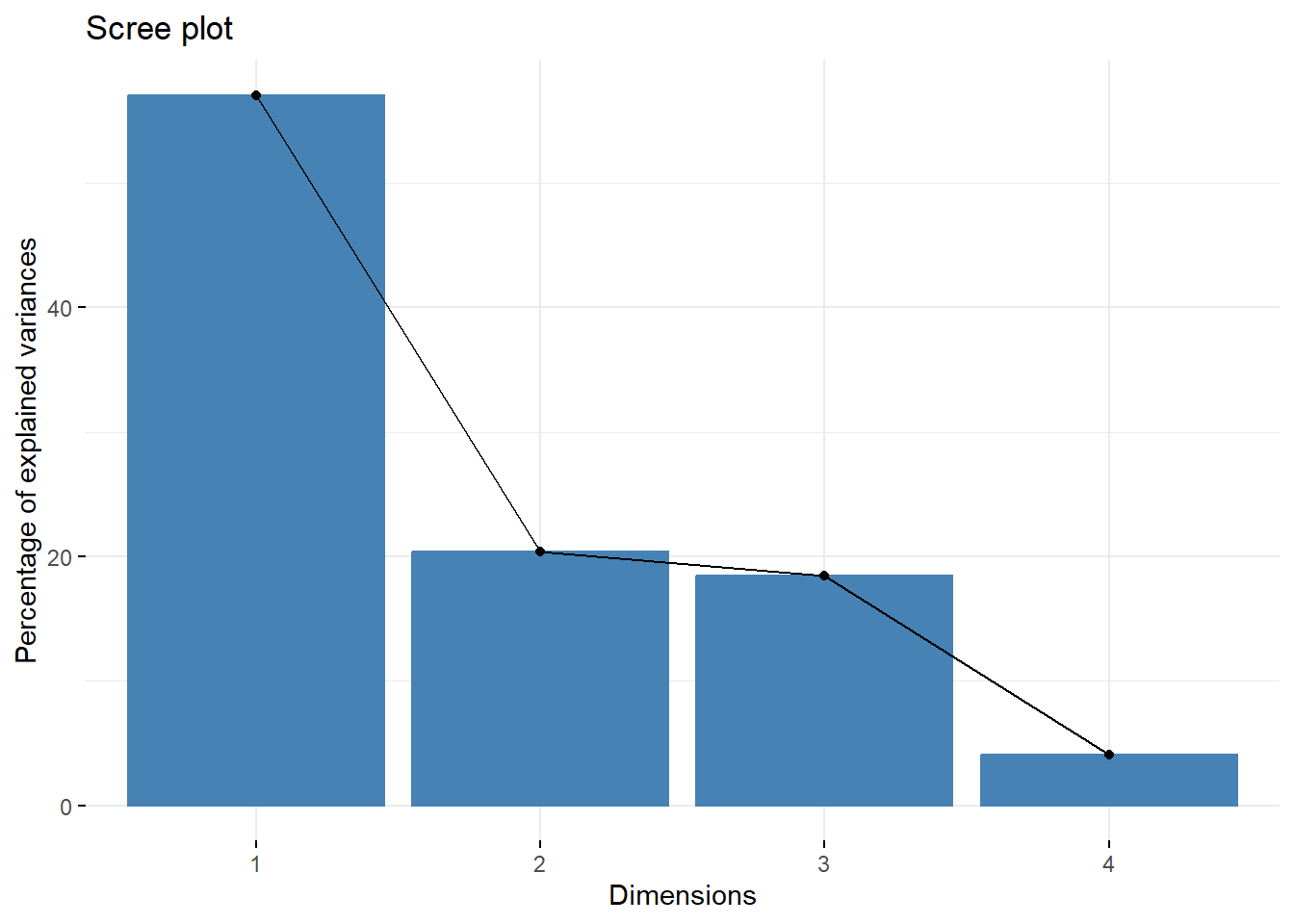
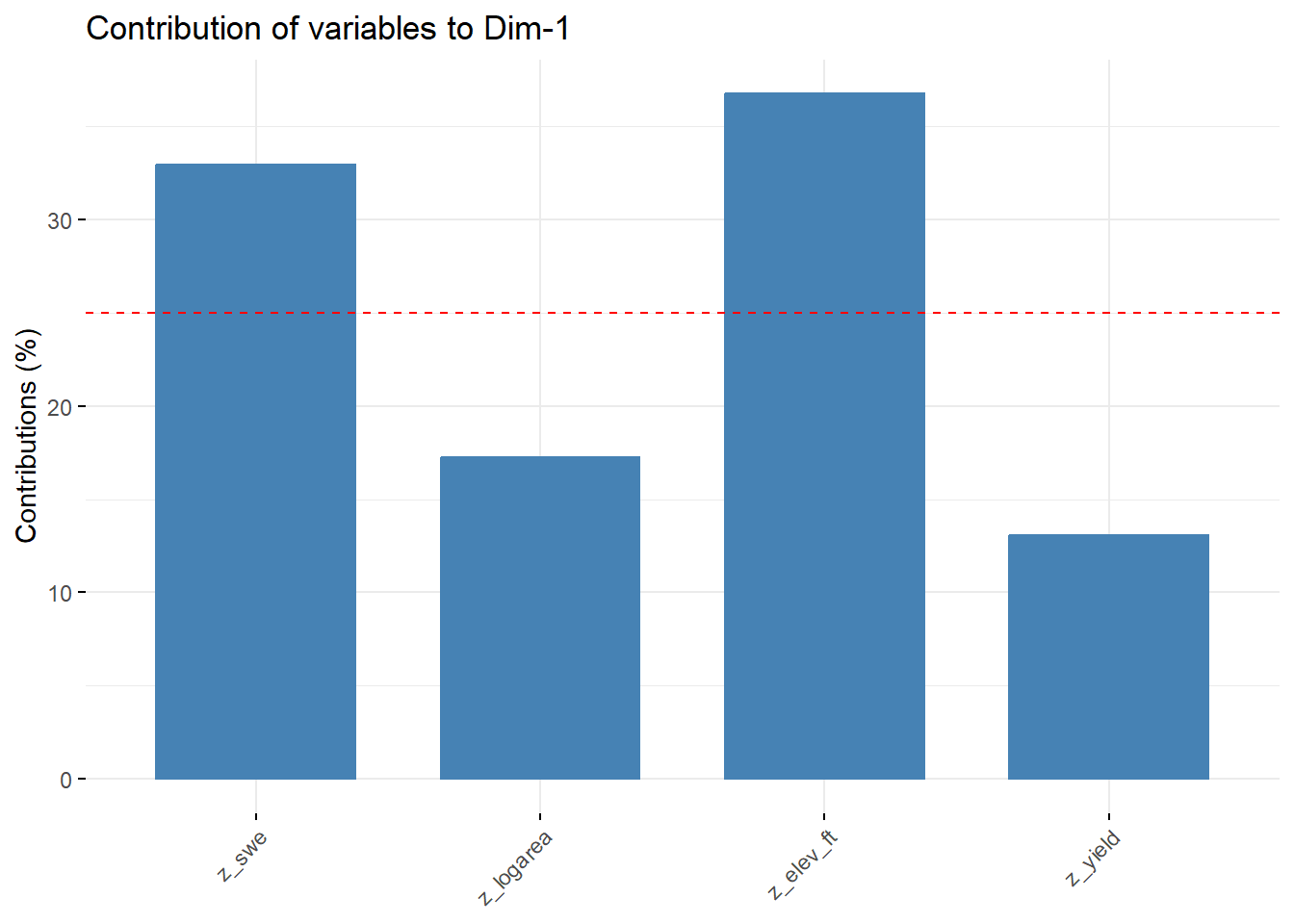
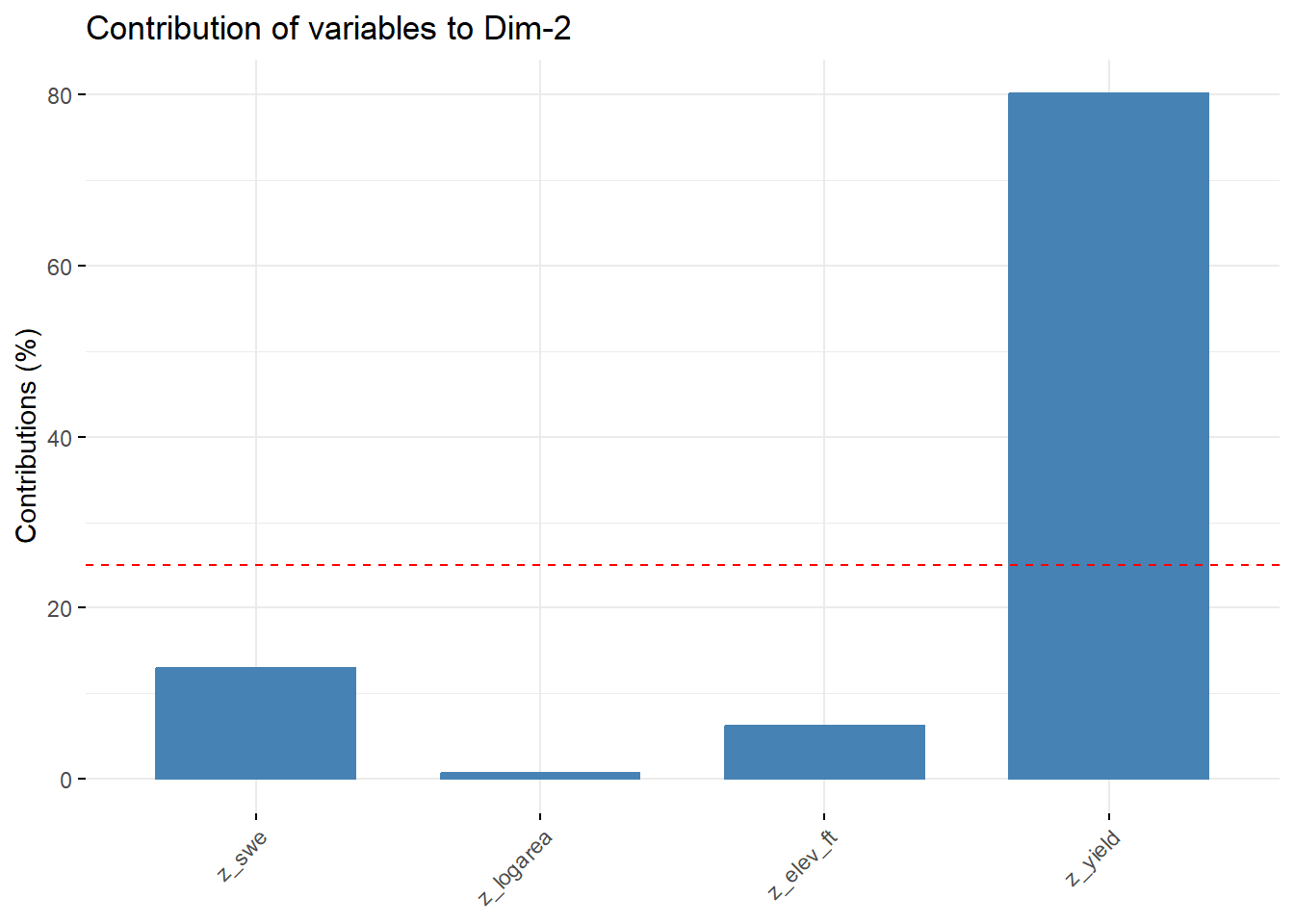
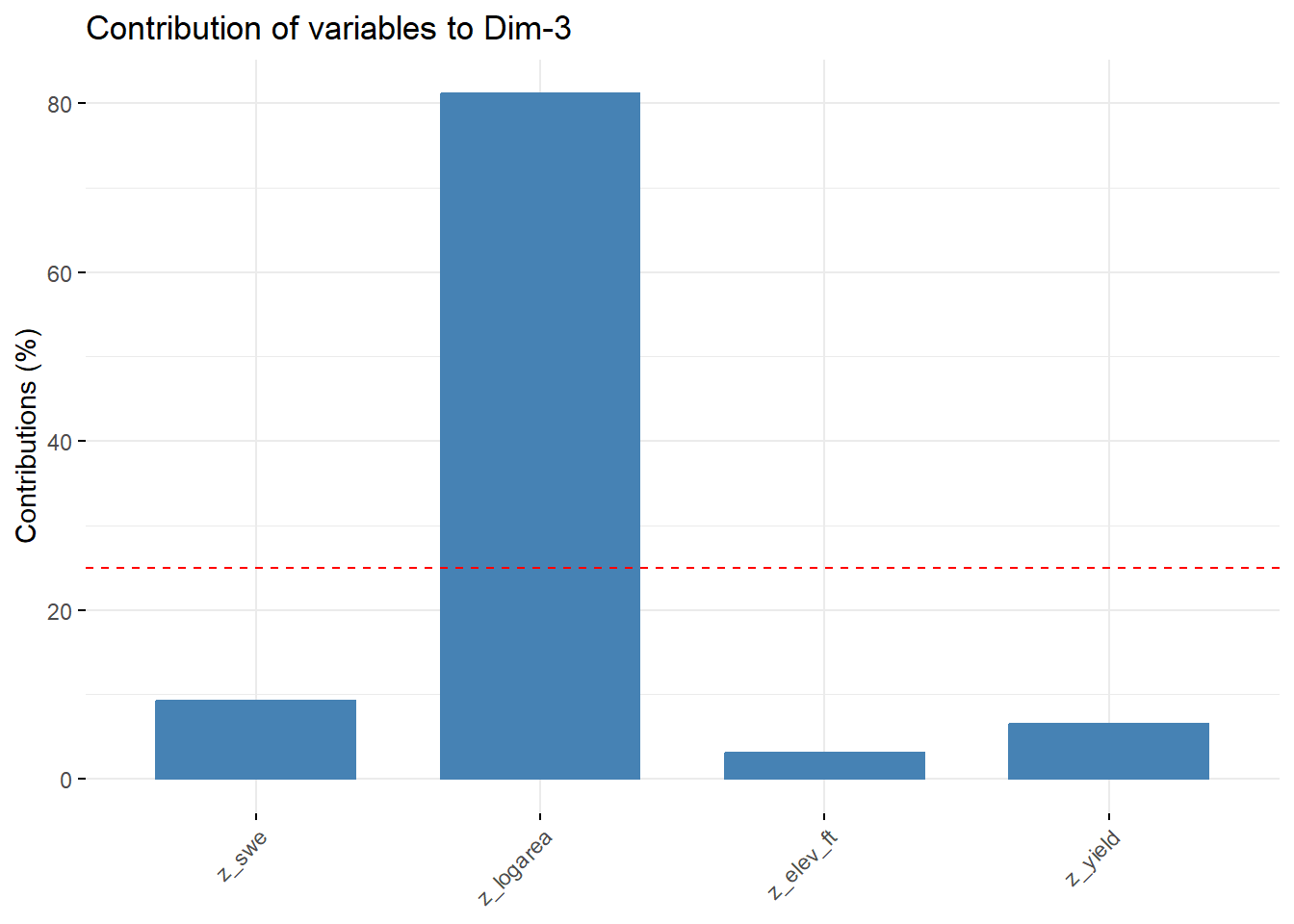
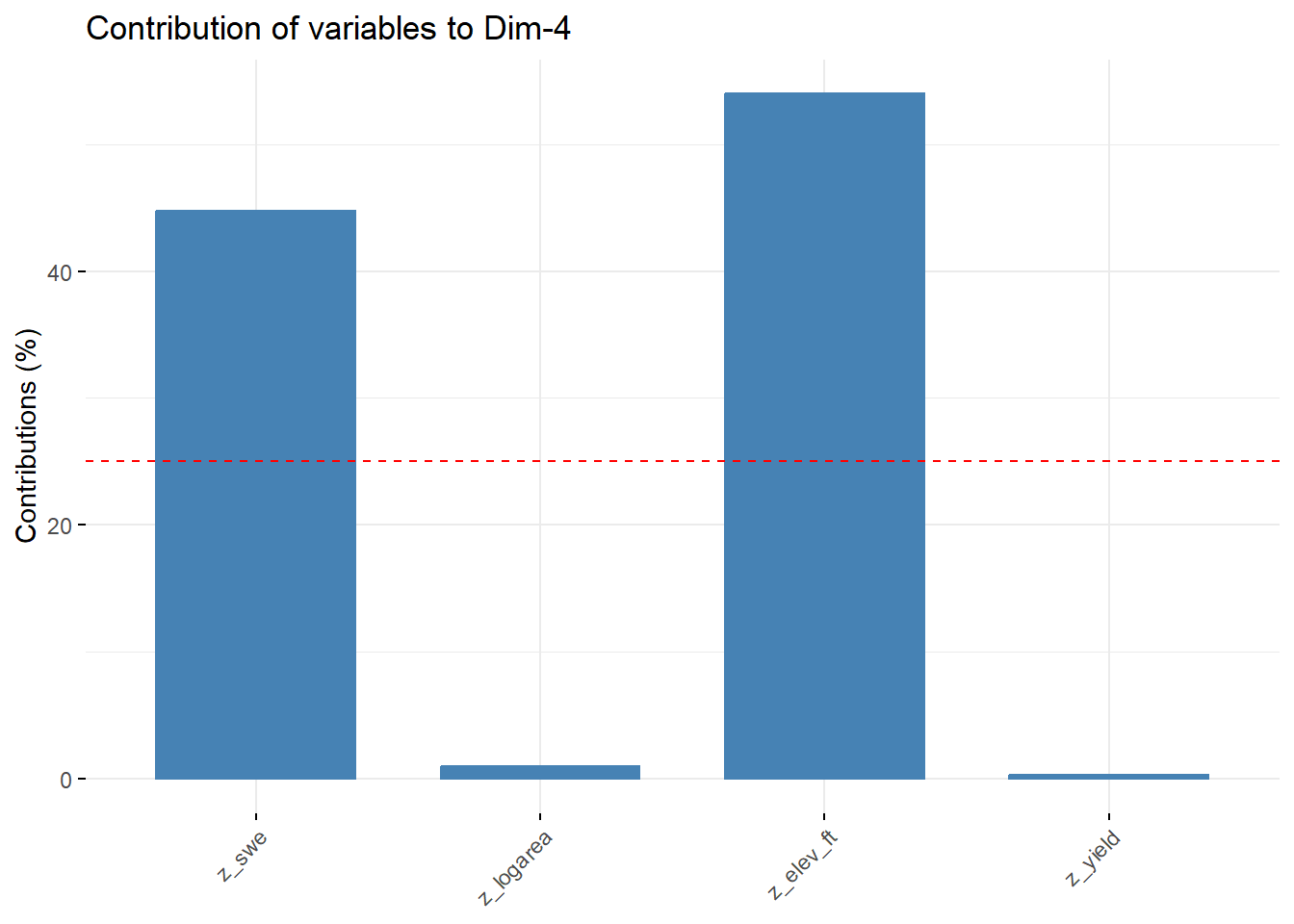
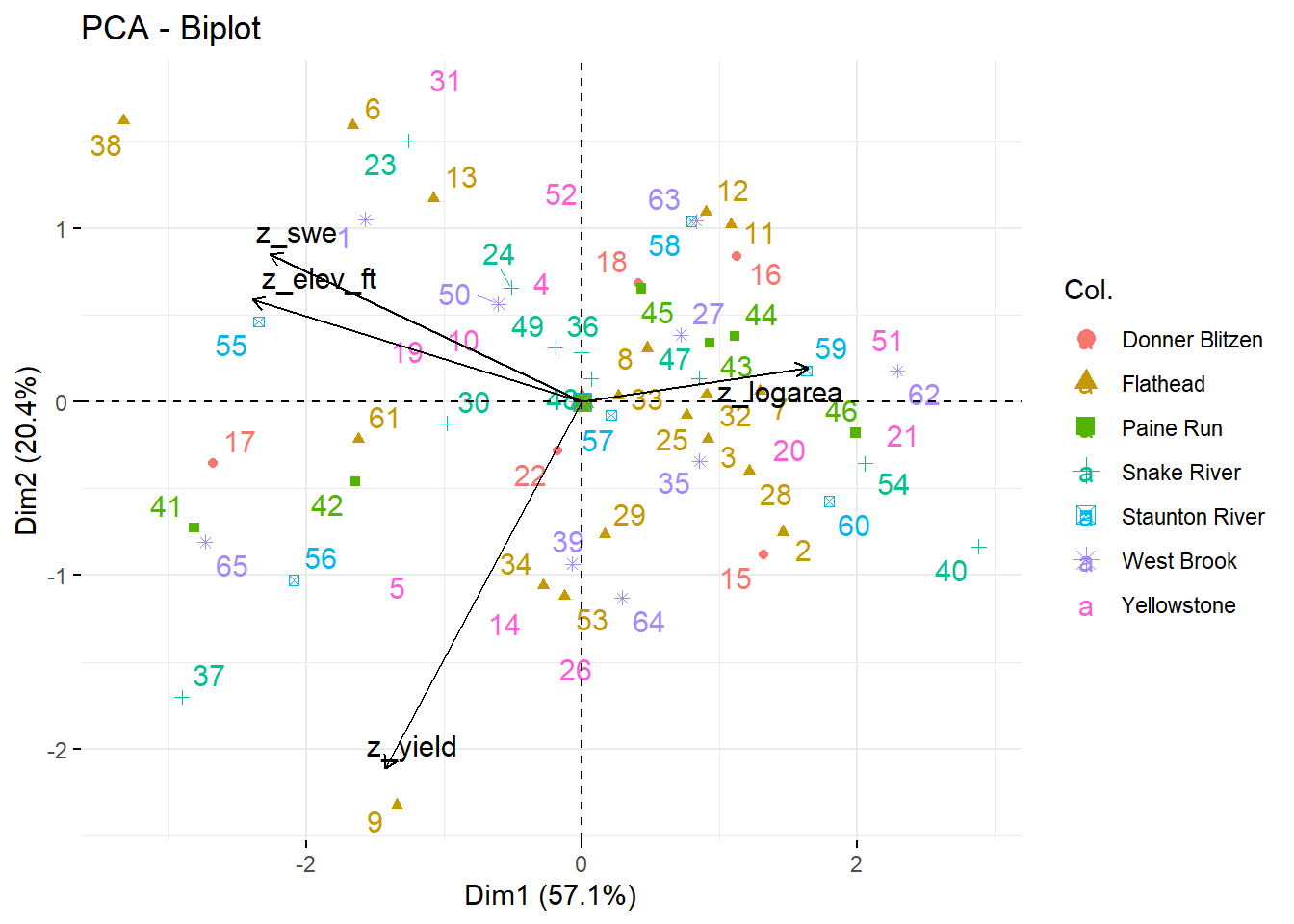
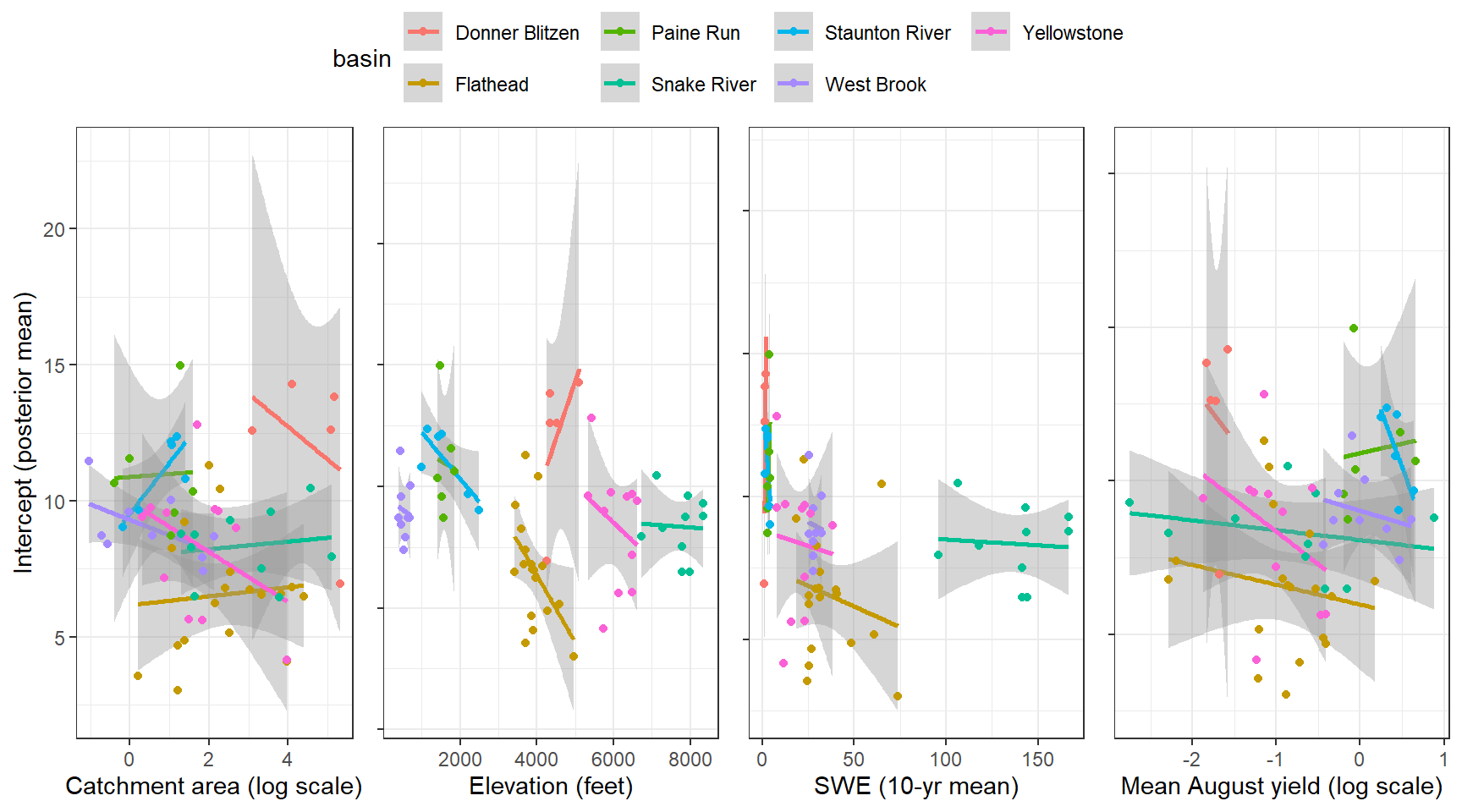
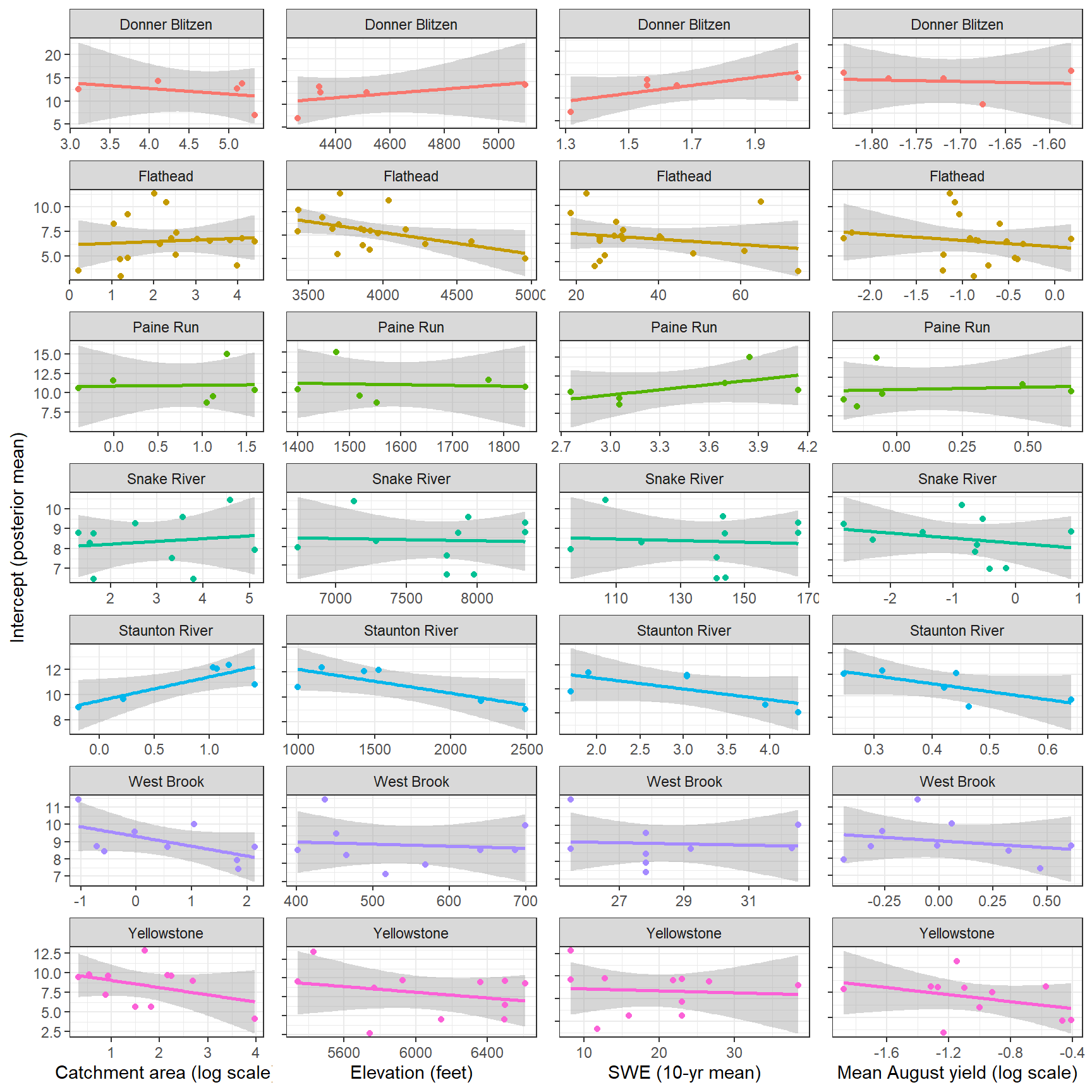
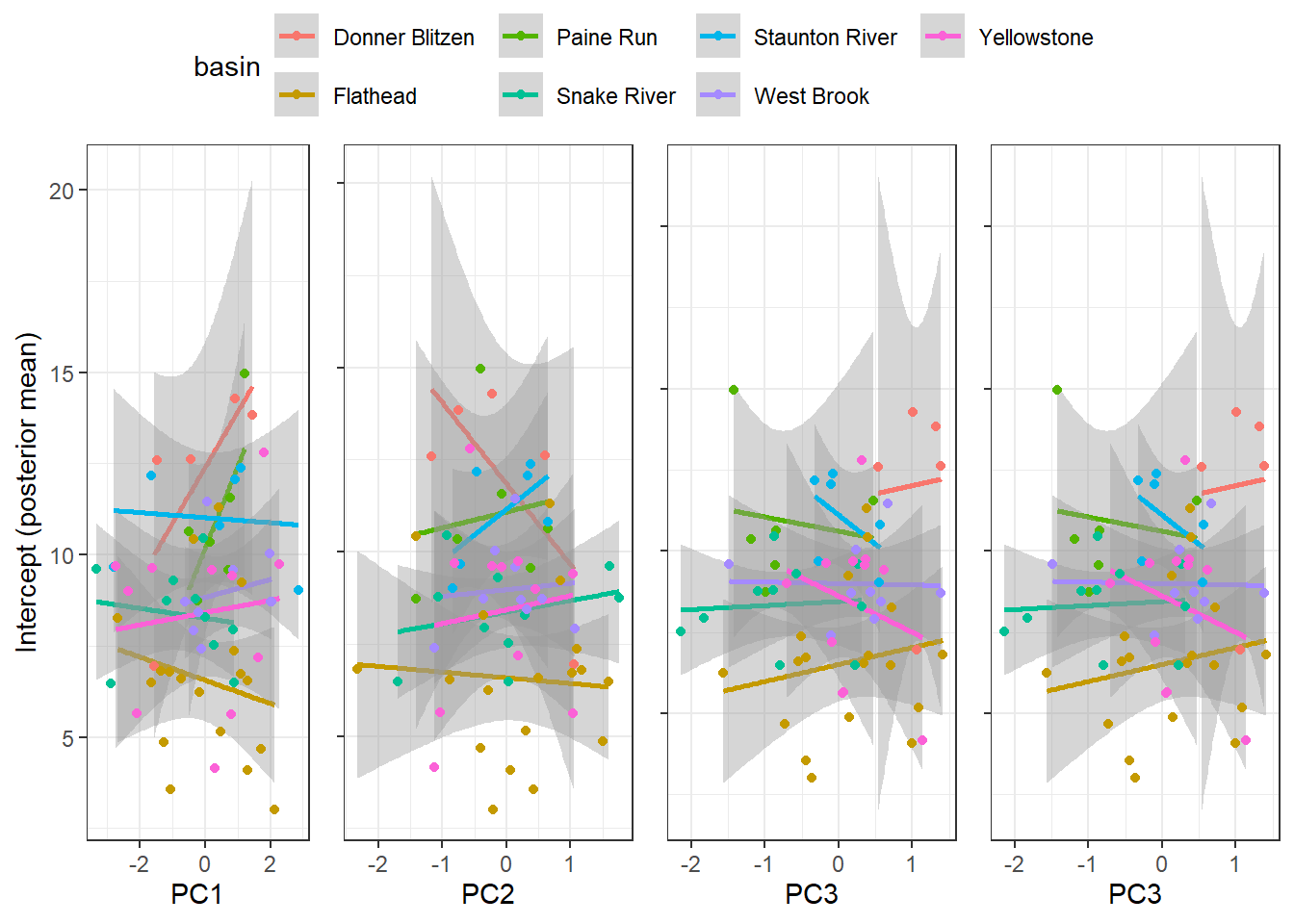
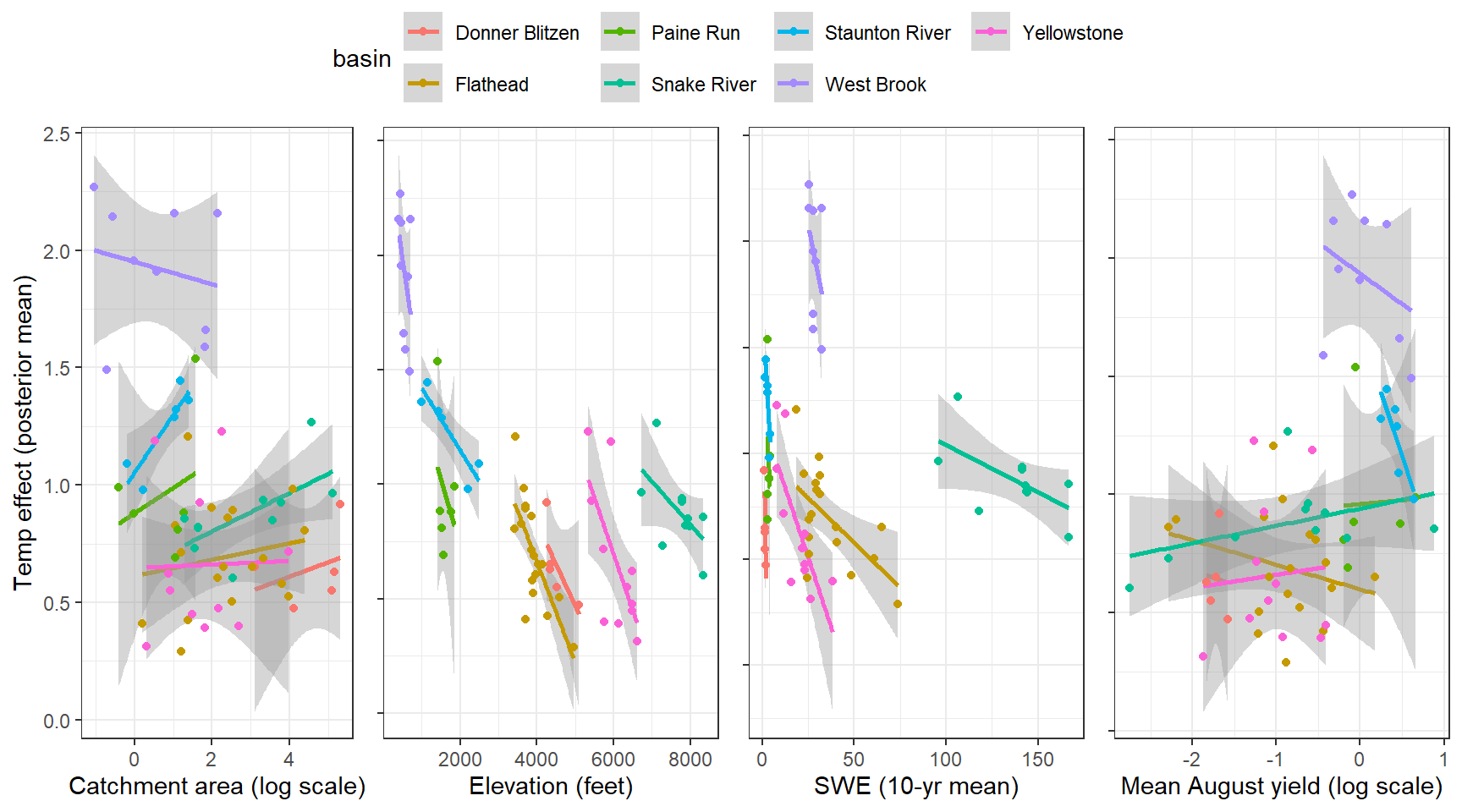
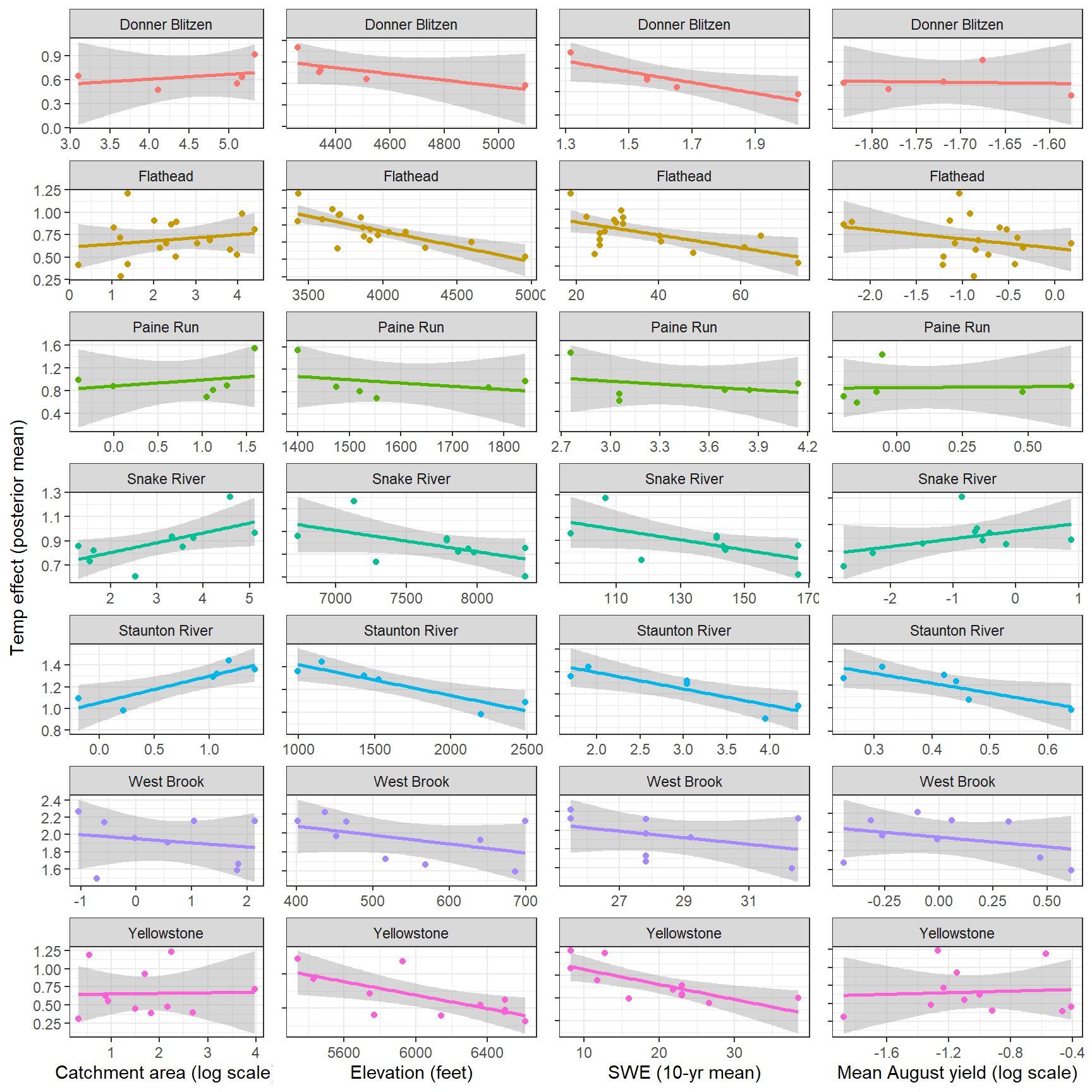
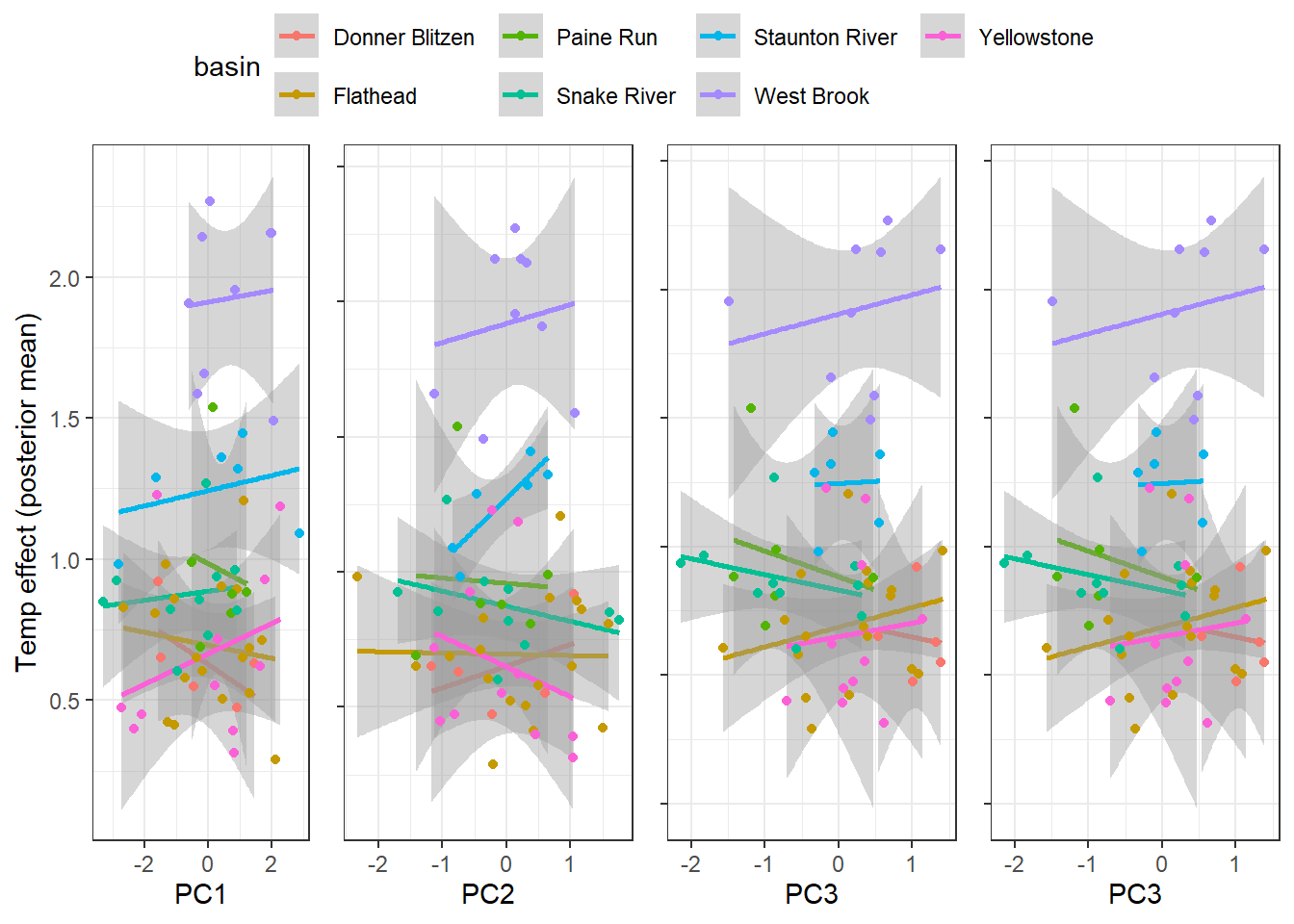
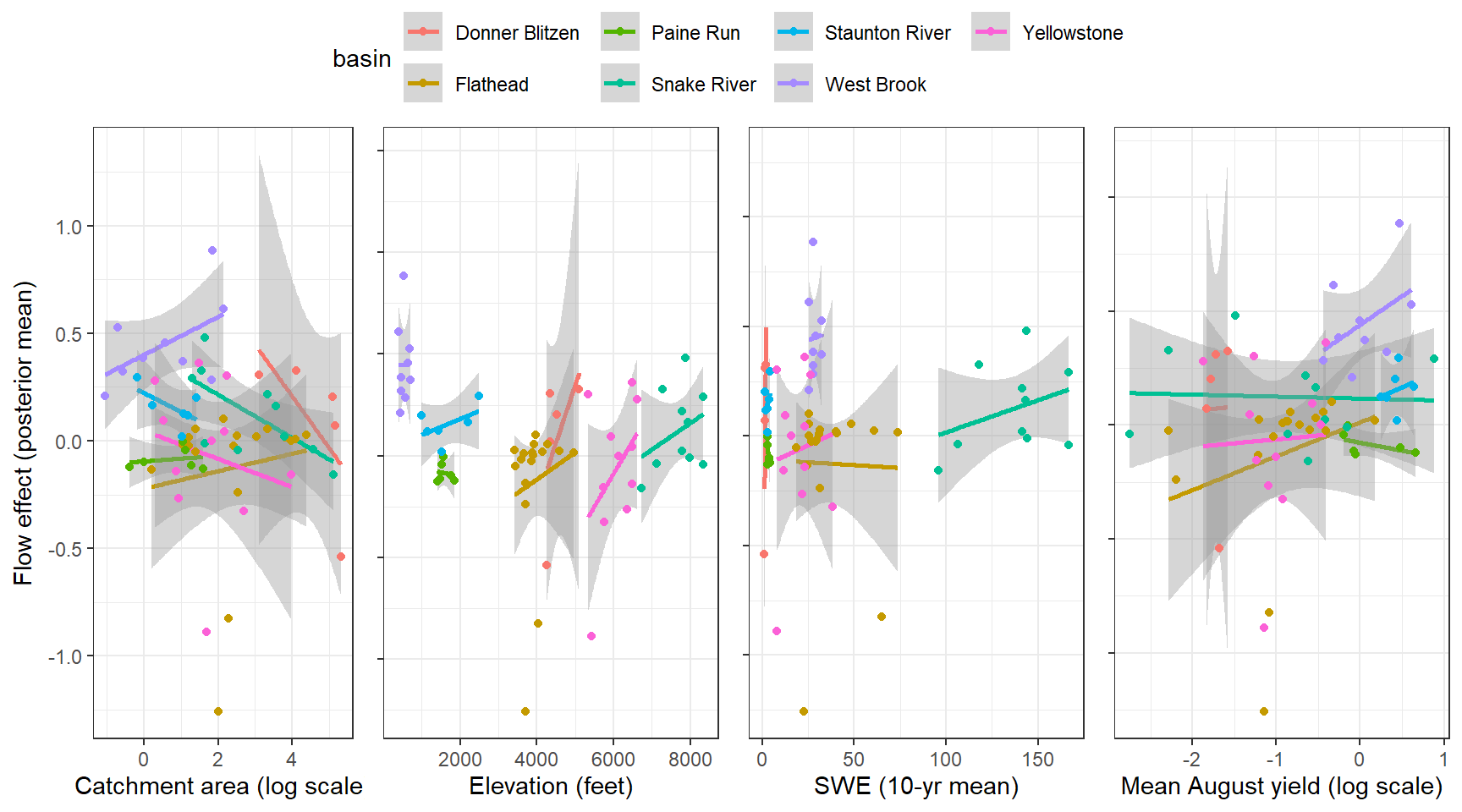
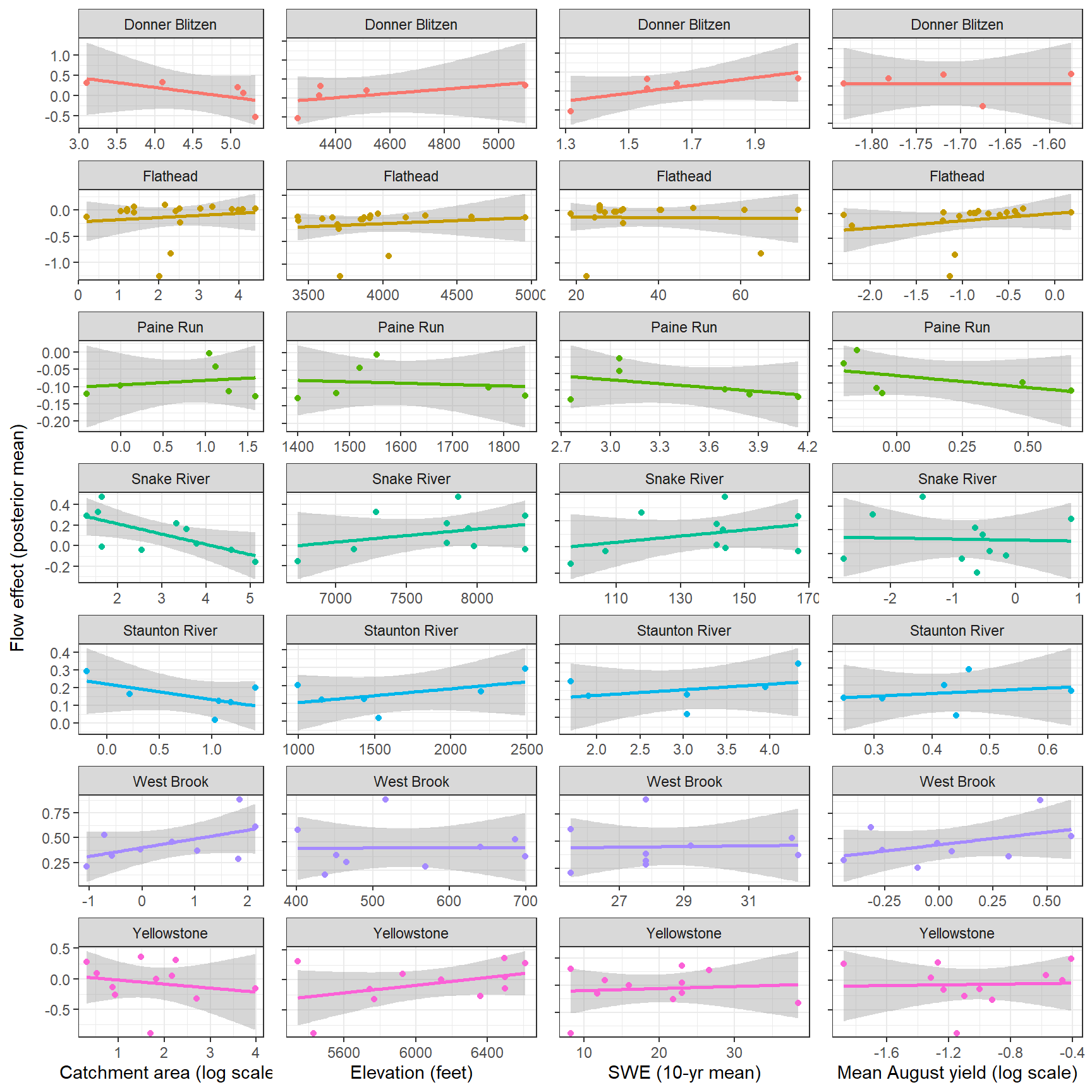
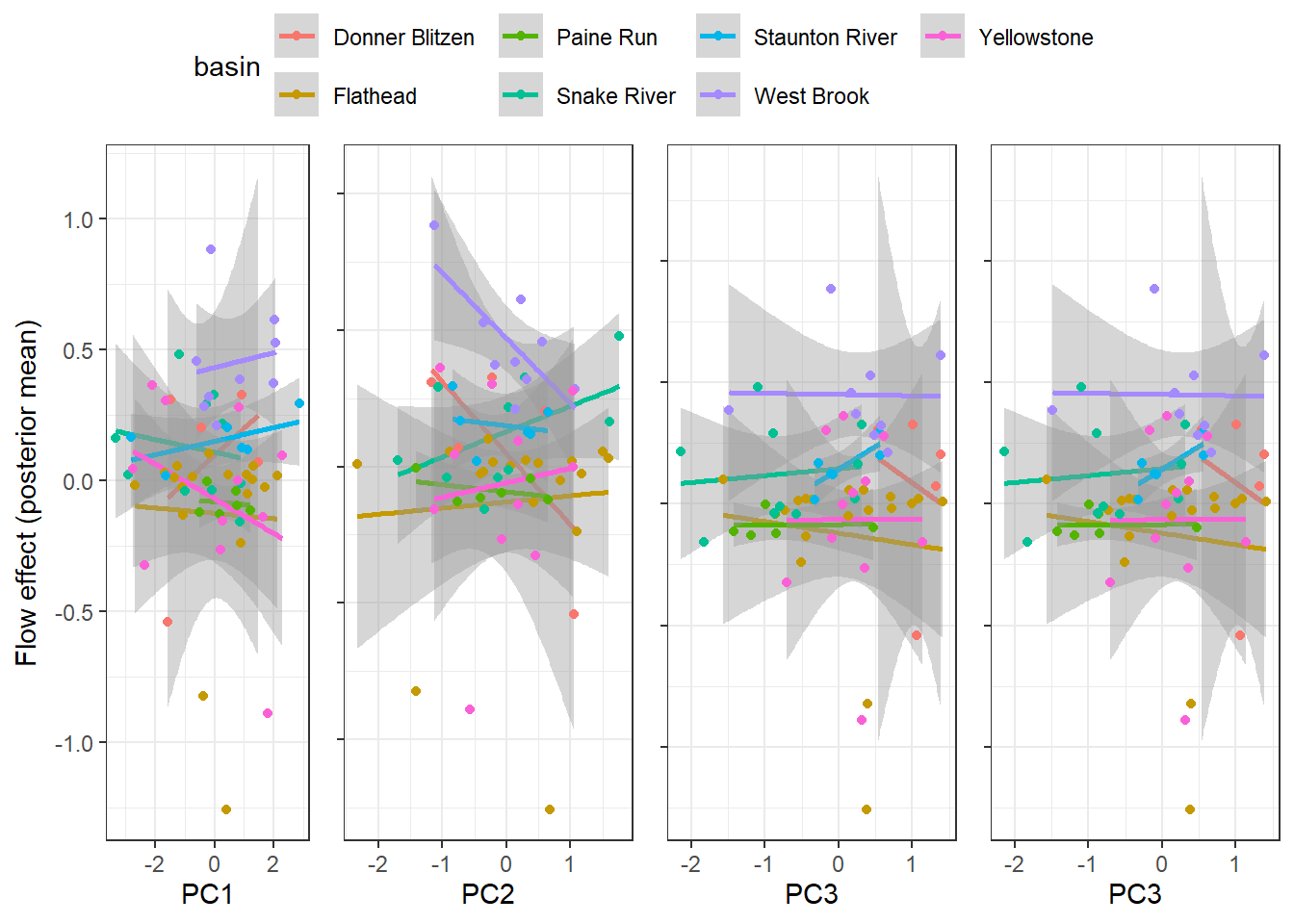
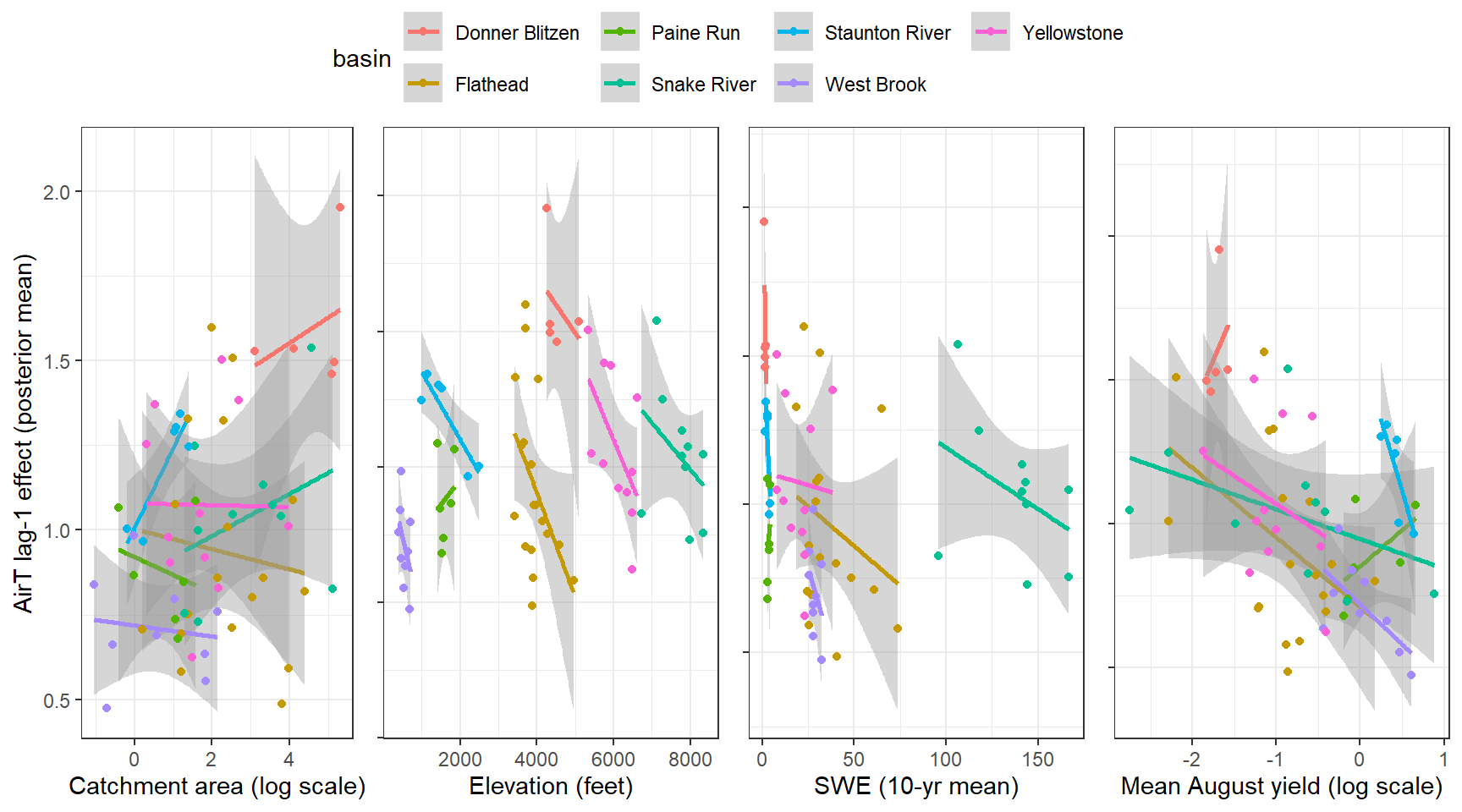
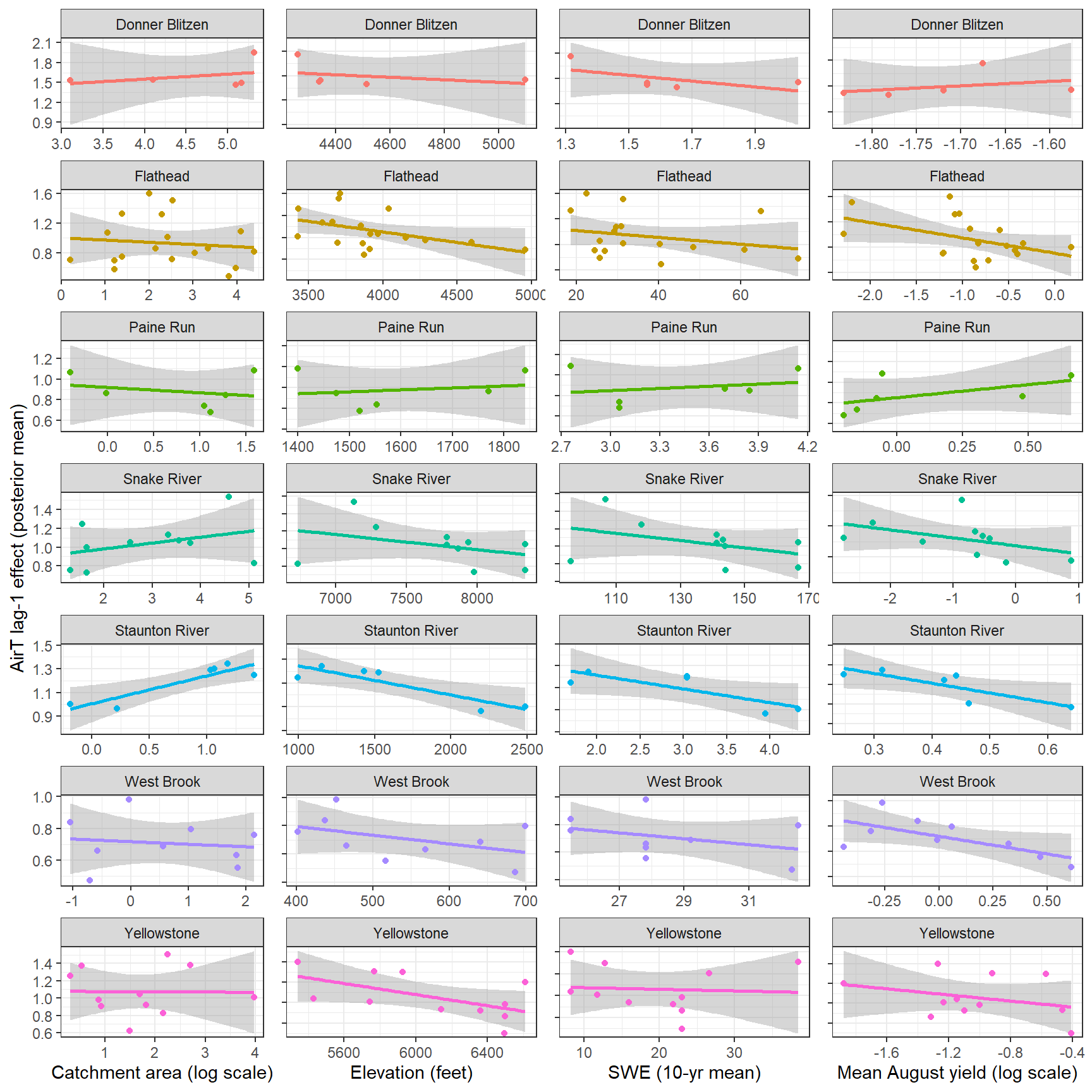
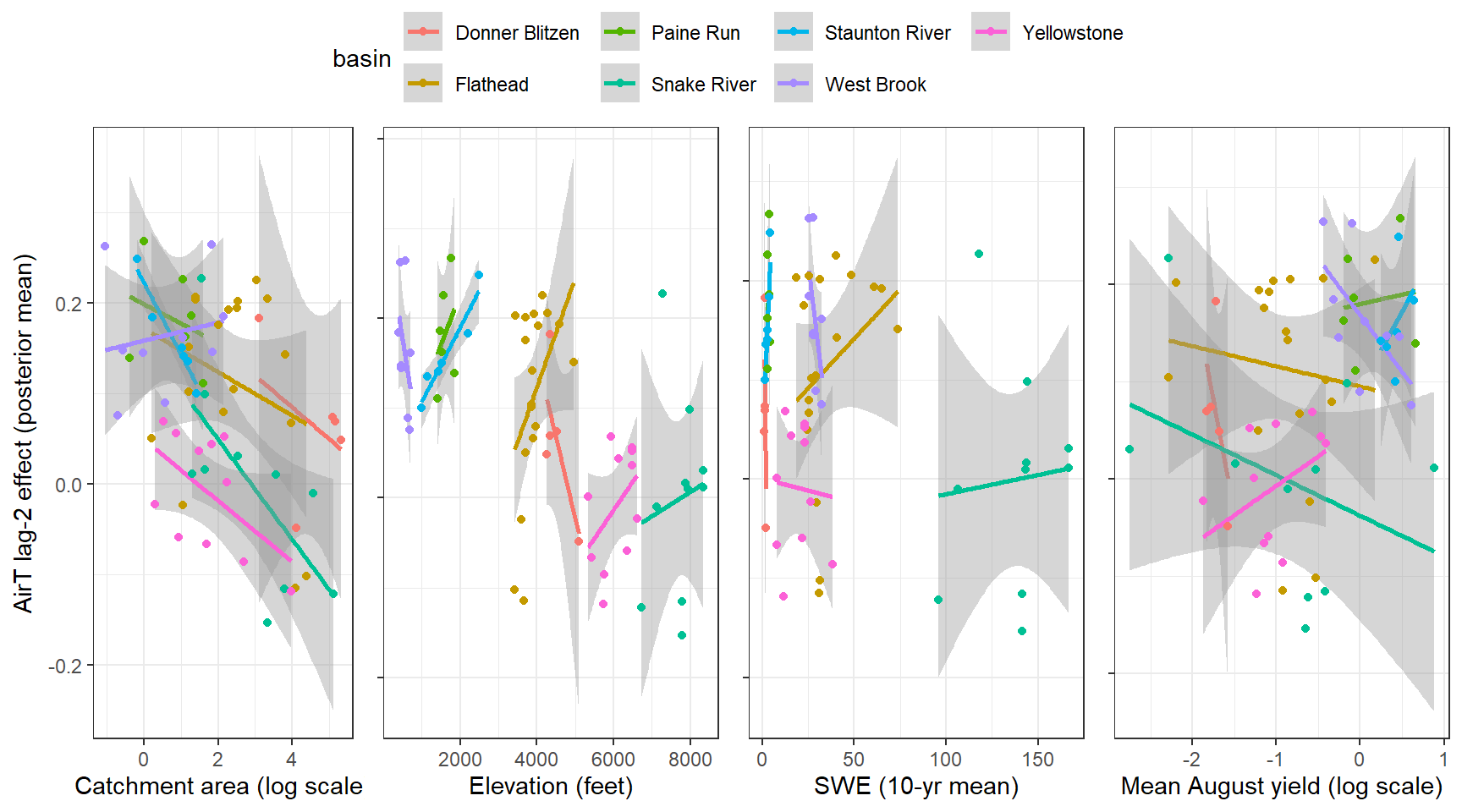
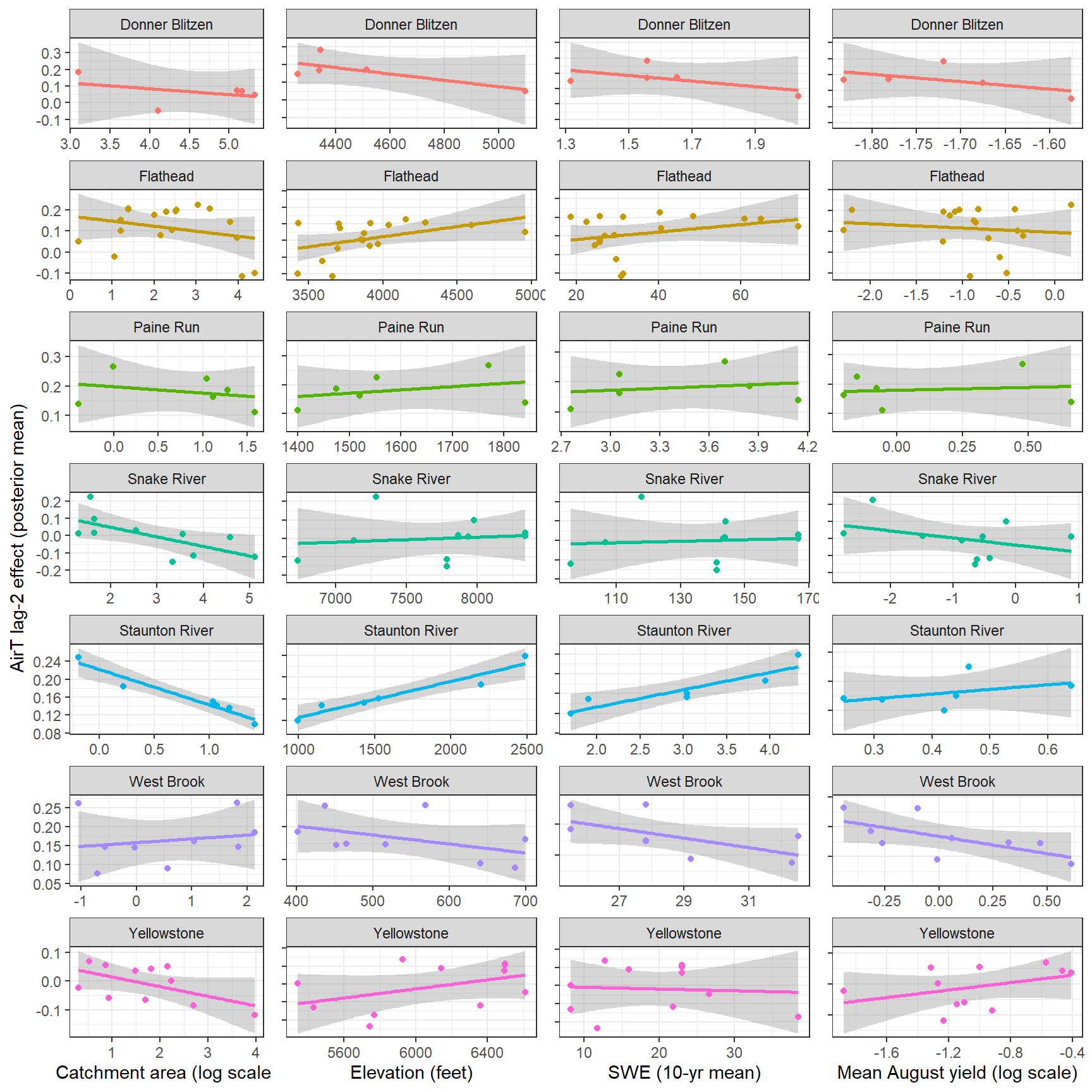
--- title: "ModelTemp: EcoD, simple" execute: cache: false --- ```{r include=FALSE} library (tidyverse)library (R2jags)library (MCMCvis)library (loo)library (HDInterval)library (scales)library (ggmcmc)library (GGally)library (beepr)library (lme4)library (factoextra)``` * Consider bringing in additional data from NWIS to fill in middle-upper end of catchment size spectrum (Snake: Blackrock, Cache, Granite...bigger rivers? Hoback, Gros Ventre, Greys...where to draw the line?)* consider putting a prior on flow variables in the JAGS model to deal with missing values (rather than setting NAs to 0, as in line 57)## Load data ```{r} <- read_csv ("data/EcoDrought_FlowTempData_formatted.csv" ) #%>% # filter(!is.na(tempc_mean)) %>% # mutate(rowNum = 1:nrow(.), # # site_code = as.numeric(as.factor(site_name)), # year_code = year - min(year) + 1) # redo basin and site codes # dat <- dat %>% arrange(basin, site_name, year, yday) # sitecodes <- tibble(site_name = unique(dat$site_name), site_code = 1:length(unique(dat$site_name))) # basincodes <- tibble(basin = unique(dat$basin), basin_code = 1:length(unique(dat$basin))) # dat <- dat %>% # select(-c(site_code, basin_code)) %>% # left_join(sitecodes) %>% # # left_join(basincodes) %>% # mutate(rowNum = 1:nrow(.)) ``` ```{r} # dropsites <- unlist(dat %>% # group_by(site_name) %>% # summarize(numt = sum(!is.na(tempc_mean)), # numf = sum(!is.na(z_Yield_mm_log))) %>% # ungroup() %>% # filter(numt == 0 | numf == 0) %>% # select(site_name)) # # dat <- dat %>% # filter(!site_name %in% dropsites) %>% # mutate(rowNum = 1:nrow(.), # site_code = as.numeric(as.factor(site_name)), # year_code = year - min(year) + 1) ``` ```{r} :: datatable (dat %>% group_by (basin, site_name, site_code) %>% summarise (n = n ()) )``` ```{r} sum (is.na (dat$ z_air_temp_mean))sum (is.na (dat$ z_air_temp_mean_lag1))sum (is.na (dat$ z_air_temp_mean_lag2))sum (is.na (dat$ z_Yield_mm_log))``` ```{r} # dat <- dat %>% mutate(z_air_temp_mean = ifelse(is.na(z_air_temp_mean), 0, z_air_temp_mean), # z_air_temp_mean_lag1 = ifelse(is.na(z_air_temp_mean_lag1), 0, z_air_temp_mean_lag1), # z_air_temp_mean_lag2 = ifelse(is.na(z_air_temp_mean_lag2), 0, z_air_temp_mean_lag2), # z_Yield_mm_log = ifelse(is.na(z_Yield_mm_log), 0, z_Yield_mm_log)) ``` #### Distributions ```{r eval=FALSE} :: ggarrange (dat %>% ggplot (aes (x = air_temp_mean, color = site_name)) + geom_density () + theme_bw (),%>% ggplot (aes (x = flow_mean_log, color = site_name)) + geom_density () + theme_bw (),%>% ggplot (aes (x = Yield_mm_log, color = site_name)) + geom_density () + theme_bw (),common.legend = TRUE , legend = "right" , ncol = 1 )``` #### Air temp ```{r eval=FALSE} ggplot (dat, aes (yday, z_air_temp_mean))+ geom_line (aes (color = factor (year))) + facet_grid (year ~ site_name)``` #### Water temp ```{r eval=FALSE} ggplot (dat, aes (yday, tempc_mean))+ geom_line (aes (color = factor (year)))+ facet_grid (year ~ site_name)``` #### Flow (log and std) ```{r eval=FALSE} ggplot (dat, aes (yday, z_Yield_mm_log))+ geom_line (aes (color = factor (year)))+ facet_grid (year ~ site_name)``` #### Tw ~ Ta + F ```{r eval=FALSE} %>% ggplot (aes (x = z_air_temp_mean, y = tempc_mean, color = z_Yield_mm_log)) + geom_point (size = 0.2 ) + facet_wrap (~ site_name) + theme_bw ()``` #### Ta ~ F ```{r eval=FALSE} %>% ggplot (aes (x = z_air_temp_mean, y = z_Yield_mm_log, colour = tempc_mean)) + geom_point (size = 0.2 ) + facet_wrap (~ site_name) + theme_bw () + ggpubr:: stat_cor ()``` #### Ta ~ yday ```{r eval=FALSE} %>% ggplot (aes (x = z_yday, y = z_air_temp_mean, colour = tempc_mean)) + geom_point (size = 0.2 ) + facet_wrap (~ site_name) + theme_bw () + ggpubr:: stat_cor ()``` #### F ~ yday ```{r eval=FALSE} %>% filter (z_Yield_mm_log != 0 ) %>% ggplot (aes (x = z_yday, y = z_Yield_mm_log, colour = tempc_mean)) + geom_point (size = 0.2 ) + facet_wrap (~ site_name) + theme_bw () + ggpubr:: stat_cor ()``` ```{r} ggpairs (dat %>% select (yday, z_air_temp_mean, z_Yield_mm_log))``` ## Specify JAGS model * add prior on covariates to deal with missing values* calculate Bayesian R^2 and RMSE* remove the year trends (same as for Hocking et al 2018)* add catchment covariate effects on random effects parameters```{r} cat ("model { ###----------------- LIKELIHOOD -----------------### # Days without an observation on the previous day (first observation in a series) # No autoregressive term for (i in 1:nFirstObsRows){ temp[firstObsRows[i]] ~ dnorm(stream.mu[firstObsRows[i]], pow(sigma, -2)) stream.mu[firstObsRows[i]] <- trend[firstObsRows[i]] trend[firstObsRows[i]] <- #inprod(B.0[], X.0[firstObsRows[i], ]) + inprod(B.site[site[firstObsRows[i]], ], X.site[firstObsRows[i], ]) #+ #inprod(B.year[year[firstObsRows[i]], ], X.year[firstObsRows[i], ]) } # Days with an observation on the previous dat (all days following the first day) # Includes autoregressive term (ar1) for (i in 1:nEvalRows){ temp[evalRows[i]] ~ dnorm(stream.mu[evalRows[i]], pow(sigma, -2)) stream.mu[evalRows[i]] <- trend[evalRows[i]] + ar1[site[evalRows[i]]] * (temp[evalRows[i]-1] - trend[ evalRows[i]-1 ]) trend[evalRows[i]] <- #inprod(B.0[], X.0[evalRows[i], ]) + inprod(B.site[site[evalRows[i]], ], X.site[evalRows[i], ]) #+ #inprod(B.year[year[evalRows[i]], ], X.year[evalRows[i], ]) } # Deal with missing data for (i in 1:n){ for (k in 1:Krandom) { X.site[i,k] ~ dnorm(0, pow(1, -2)) } } # Save log-likelihoods for LOO-CV for (i in 1:n) { loglik[i] <- logdensity.norm(temp[i], stream.mu[i], pow(sigma, -2)) } ###----------------- PRIORS ---------------------### # ar1, hierarchical by site for (i in 1:nsites){ ar1[i] ~ dnorm(ar1Mean, pow(ar1SD, -2)) T(-1,1) } ar1Mean ~ dunif(-1, 1) ar1SD ~ dunif(0, 2) # model variance sigma ~ dunif(0, 100) # fixed effect coefficients # for (k in 1:Kfixed) { # B.0[k] ~ dnorm(0, 0.001) # } # random effect coefficients (by site) for (k in 1:Krandom) { sigma.B.site[k] ~ dunif(0, 100) #alpha.0[k] ~ dnorm(0, pow(10, -2)) #alpha.1[k] ~ dnorm(0, pow(10, -2)) for (i in 1:nsites) { B.site[i,k] ~ dnorm(0, pow(sigma.B.site[k], -2)) #mu.B.site[i,k] <- alpha.0[k] + alpha.1[k] * area[i] } } # YEAR EFFECTS # for(l in 1:L) { # sigma.b.year[l] ~ dunif(0, 100) # for(t in 1:Ti){ # B.year[t, l] ~ dnorm(0, pow(sigma.b.year[l], -2)) # } # } # # YEAR EFFECTS # # Priors for random effects of year # for (t in 1:Ti) { # Ti years # B.year[t, 1:L] ~ dmnorm(mu.year[ ], tau.B.year[ , ]) # } # # mu.year[1] <- 0 # # for (l in 2:L) { # mu.year[l] ~ dnorm(0, 0.0001) # } # # # Prior on multivariate normal std deviation # tau.B.year[1:L, 1:L] ~ dwish(W.year[ , ], df.year) # df.year <- L + 1 # sigma.B.year[1:L, 1:L] <- inverse(tau.B.year[ , ]) # for (l in 1:L) { # for (l.prime in 1:L) { # rho.B.year[l, l.prime] <- sigma.B.year[l, l.prime]/sqrt(sigma.B.year[l, l]*sigma.B.year[l.prime, l.prime]) # } # sigma.b.year[l] <- sqrt(sigma.B.year[l, l]) # } ###----------------- DERIVED VALUES -------------### #residuals[1] <- 0 # hold the place. Not sure if this is necessary... for (i in 1:n) { residuals[i] <- temp[i] - stream.mu[i] } # variance of model predictions (fixed + random effects) var_fit <- (sd(stream.mu))^2 # residual variance var_res <- (sd(residuals))^2 # calculate Bayesian R^2 R2 <- var_fit / (var_fit + var_res) # Root mean squared error rmse <- sqrt(mean(residuals[]^2)) }" , file = "JAGS models/DailyTempModelJAGS_Letcher_hierarchical_modified.txt" )``` ## Organize objects ```{r} # row indices for first observation in each site-year <- unlist (dat %>% group_by (siteYear) %>% summarize (index = rowNum[min (which (! is.na (tempc_mean)))]) %>% ungroup () %>% select (index))<- length (firstObsRows)# does the number of first observations match the number of site years? == length (unique (dat$ siteYear))``` ```{r} <- unlist (dat %>% filter (! rowNum %in% firstObsRows) %>% select (rowNum))<- length (evalRows)``` ```{r} <- data.frame (airTempLag1 = dat$ z_air_temp_mean_lag1,airTempLag2 = dat$ z_air_temp_mean_lag2) <- data.frame (intercept = 1 ,airTemp = dat$ z_air_temp_mean, flow = dat$ z_Yield_mm_log,airFlow = dat$ z_air_temp_mean * dat$ z_Yield_mm_log,airTempLag1 = dat$ z_air_temp_mean_lag1,airTempLag2 = dat$ z_air_temp_mean_lag2# doy = dat$z_yday, # doyFlow = dat$z_yday * dat$z_Yield_mm_log <- data.frame (intercept.year = rep (1 , length.out = dim (data.fixed)[1 ])#, #dOY = dat$yday, #dOY2 = dat$yday^2, #dOY3 = dat$yday^3 <- log (dat %>% group_by (site_code) %>% summarize (area_sqmi = unique (area_sqmi)) %>% ungroup () %>% arrange (site_code) %>% pull (area_sqmi))any (is.na (data.random))any (is.na (data.fixed))any (is.na (data.random.years))any (is.na (data.random.covs))``` ```{r} <- length (unique (dat$ year))<- dim (data.random.years)[2 ]<- diag (L)``` ```{r} # combine data in a list <- list ("temp" = dat$ tempc_mean,"nFirstObsRows" = nFirstObsRows,"firstObsRows" = firstObsRows,"nEvalRows" = nEvalRows,"evalRows" = evalRows,"X.0" = as.matrix (data.fixed),"X.site" = as.matrix (data.random),"X.year" = as.matrix (data.random.years),"Kfixed" = dim (data.fixed)[2 ],"Krandom" = dim (data.random)[2 ],"nsites" = length (unique (dat$ site_name)),"Ti" = Ti,"L" = L,"W.year" = W.year,"n" = dim (dat)[1 ],"year" = dat$ year_code,"site" = dat$ site_code,"area" = data.random.covs``` ```{r} <- c ("residuals" ,"deviance" ,"sigma" ,"B.0" ,"B.site" ,"B.year" ,"rho.B.year" ,"mu.year" ,"sigma.b.year" ,"stream.mu" ,"ar1" ,"ar1Mean" ,"ar1SD" ,"temp" ,"sigma.B.site" ,"R2" ,"rmse" ,"alpha.0" ,"alpha.1" ,"loglik" ``` ## Fit model ```{r eval=FALSE} <- Sys.time ()<- jags.parallel (data = jags.data, inits = NULL , parameters.to.save = jags.params, model.file = "JAGS models/DailyTempModelJAGS_Letcher_hierarchical_modified.txt" ,n.chains = 15 , n.thin = 20 , n.burnin = 1000 , n.iter = 5000 , DIC = TRUE )Sys.time () - st``` #### Save model ouput ```{r eval=FALSE} saveRDS (fit_ed, "Model objects/LetcherTempModel_EcoDrought_hierarchical_fullish.RDS" )``` ```{r cache=FALSE} <- readRDS ("Model objects/LetcherTempModel_EcoDrought_hierarchical_fullish.RDS" )``` ```{r cache=FALSE} <- fit_ed# generate MCMC samples and store as an array <- top_mod$ BUGSoutput<- vector ("list" , length = dim (modelout$ sims.array)[2 ])for (i in 1 : length (McmcList)) { McmcList[[i]] = coda:: as.mcmc (modelout$ sims.array[,i,]) }# rbind MCMC samples from 10 chains <- rbind (McmcList[[1 ]], McmcList[[2 ]], McmcList[[3 ]], McmcList[[4 ]], McmcList[[5 ]], McmcList[[6 ]], McmcList[[7 ]], McmcList[[8 ]], McmcList[[9 ]], McmcList[[10 ]], McmcList[[11 ]], McmcList[[12 ]], McmcList[[13 ]], McmcList[[14 ]], McmcList[[15 ]])<- modelout$ summaryhead (param.summary)rm (fit_ed)``` #### Check convergence ```{r} $ BUGSoutput$ summary[,8 ][top_mod$ BUGSoutput$ summary[,8 ] > 1.05 ]``` ```{r eval=FALSE} MCMCtrace (top_mod, ind = TRUE , params = c (#"B.0", "B.site" , # "B.year", # "ar1", # "alpha.0", # "alpha.1", "sigma" ), pdf = FALSE )``` ```{r cache=FALSE} <- ggs (coda:: as.mcmc (top_mod), keep_original_order = TRUE )head (ggfit)``` ### LOO-CV ```{r cache=FALSE} <- top_mod$ BUGSoutput$ sims.list$ loglik # extract the log-likelihood estimates for each MCMC sample <- relative_eff (exp (loglik), chain_id = rep (1 : 15 , each = 200 ))<- loo (loglik, r_eff = rf)plot (my_loo)# badpts <- which(my_loo$diagnostics$pareto_k > 0.7) ``` ### Goodness of fit ```{r cache=FALSE} <- as_tibble (Mcmcdat)# subset expected and observed MCMC samples <- as.matrix (Mcmcdat[,startsWith (names (Mcmcdat), "stream.mu[" )])<- as.matrix (Mcmcdat[,startsWith (names (Mcmcdat), "temp[" )])``` ```{r} sum (ppdat_exp > ppdat_obs) / (dim (ppdat_obs)[1 ]* dim (ppdat_obs)[2 ])``` ```{r} <- apply (ppdat_obs, 2 , mean)<- apply (ppdat_exp, 2 , mean)tibble (obs = ppdat_obs_mean, exp = ppdat_exp_mean) %>% ggplot (aes (x = obs, y = exp)) + geom_point (alpha = 0.1 ) + # geom_smooth(method = "lm") + geom_abline (intercept = 0 , slope = 1 , color = "red" ) + theme_bw () + theme (panel.grid = element_blank ()) + xlab ("Observed" ) + ylab ("Predicted (mean)" )``` ```{r fig.height=10, fig.width=10, eval=FALSE} tibble (obs = ppdat_obs_mean, exp = ppdat_exp_mean, site = dat$ site_name, year = dat$ year) %>% ggplot (aes (x = obs, y = exp)) + geom_point (alpha = 0.1 ) + # geom_smooth(method = "lm") + geom_abline (intercept = 0 , slope = 1 , color = "red" ) + theme_bw () + theme (panel.grid = element_blank ()) + xlab ("Observed" ) + ylab ("Predicted (mean)" ) + facet_grid (year ~ site)``` ```{r} mean (unlist (ggfit %>% filter (Parameter == "rmse" ) %>% select (value)))ggs_density (ggfit, "rmse" ) + theme_bw () + theme (panel.grid = element_blank ())``` ```{r} mean (unlist (ggfit %>% filter (Parameter == "R2" ) %>% select (value)))ggs_density (ggfit, "R2" ) + theme_bw () + theme (panel.grid = element_blank ())``` ```{r} ggplot () + geom_histogram (aes (x = c (ppdat_obs_mean - ppdat_exp_mean)), color = "black" , fill = "grey" ) + xlab ("Residuals" ) + theme_bw () + theme (panel.grid = element_blank ()) + geom_vline (xintercept = 0 , color = "red" ) + xlim (- 4 ,4 )``` ```{r} tibble (site_name = dat$ site_name, year = dat$ year, yday = dat$ yday, resid = c (ppdat_obs_mean - ppdat_exp_mean)) %>% ggplot (aes (x = yday, y = resid)) + geom_point (alpha = 0.1 ) + geom_hline (yintercept = c (0 ), color = "red" ) + geom_smooth (se = FALSE , linewidth = 1.5 ) + theme_bw () + #theme(panel.grid = element_blank()) + xlab ("Day of year" ) + ylab ("Residual" ) + #facet_wrap(~site_name) + coord_cartesian (ylim = c (- 3 ,3 )) #+ facet_wrap(~year) ``` ## Plot model output ```{r} # myparams <- unique(ggfit$Parameter) <- c (dat %>% group_by (site_name) %>% summarize (basin = unique (basin)) %>% arrange (basin) %>% ungroup () %>% pull (basin))<- c (dat %>% group_by (site_name) %>% summarize (site_code = unique (site_code)) %>% arrange (site_code) %>% ungroup () %>% pull (site_name))``` ### Dot plots ##### Air temp lag 1 ```{r, fig.width=7, fig.height=7} %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",5]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",5]" , sep = "" ))) %>% ggs_caterpillar (sort = FALSE ) + aes (color = mybasins) + scale_y_discrete (labels = rev (mysites), limits = rev) + xlab ("Air temp lag 1" ) + theme_bw ()``` ##### Air temp lag 2 ```{r, fig.width=7, fig.height=7} %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",6]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",6]" , sep = "" ))) %>% ggs_caterpillar (sort = FALSE ) + aes (color = mybasins) + scale_y_discrete (labels = rev (mysites), limits = rev) + xlab ("Air temp lag 2" ) + theme_bw ()``` ##### Lag effects correlation ```{r} tibble (site_name = mysites,lag1_mean = ggfit %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",5]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",5]" , sep = "" ))) %>% group_by (Parameter) %>% summarize (lag1_mean = mean (value)) %>% ungroup () %>% pull (lag1_mean),lag2_mean = ggfit %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",6]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",6]" , sep = "" ))) %>% group_by (Parameter) %>% summarize (lag2_mean = mean (value)) %>% ungroup () %>% pull (lag2_mean),basin = mybasins) %>% ggplot (aes (x = lag1_mean, y = lag2_mean, color = basin)) + geom_point () + theme_bw ()``` ##### Site intercepts ```{r, fig.width=7, fig.height=7} %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",1]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",1]" , sep = "" ))) %>% ggs_caterpillar (sort = FALSE ) + aes (color = mybasins) + scale_y_discrete (labels = rev (mysites), limits = rev) + xlab ("Intercepts" ) + theme_bw ()``` ##### Site slopes - temp ```{r, fig.width=7, fig.height=7} %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",2]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",2]" , sep = "" ))) %>% ggs_caterpillar (sort = FALSE ) + aes (color = mybasins) + scale_y_discrete (labels = rev (mysites), limits = rev) + xlab ("Slopes, temperature effect" ) + theme_bw ()``` ##### Site slopes - flow ```{r, fig.width=7, fig.height=7} %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",3]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",3]" , sep = "" ))) %>% ggs_caterpillar (sort = FALSE ) + aes (color = mybasins) + scale_y_discrete (labels = rev (mysites), limits = rev) + xlab ("Slopes, flow effect" ) + theme_bw () + geom_vline (xintercept = 0 , linetype = "dashed" )``` ##### Site slopes - temp x flow ```{r, fig.width=7, fig.height=7} %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",4]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",4]" , sep = "" ))) %>% ggs_caterpillar (sort = FALSE ) + aes (color = mybasins) + scale_y_discrete (labels = rev (mysites), limits = rev) + xlab ("Slopes, temp-flow interaction" ) + theme_bw () + geom_vline (xintercept = 0 , linetype = "dashed" )``` ##### Autoregressive terms ```{r, fig.width=7, fig.height=7} ggs_caterpillar (ggfit %>% filter (Parameter %in% paste ("ar1[" , c (1 : jags.data$ nsites), "]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("ar1[" , c (1 : jags.data$ nsites), "]" , sep = "" ))),sort = FALSE ) + scale_y_discrete (labels = rev (mysites), limits = rev) + theme_bw () + aes (color = mybasins) + xlab ("Autoregressive parameter" )``` ##### Year effects (offsets) ```{r eval=FALSE} %>% filter (Parameter %in% c ("B.year[1,1]" , "B.year[2,1]" , "B.year[3,1]" , "B.year[4,1]" , "B.year[5,1]" ,"B.year[6,1]" , "B.year[7,1]" , "B.year[8,1]" , "B.year[9,1]" , "B.year[10,1]" , "B.year[11,1]" , "B.year[12,1]" , "B.year[13,1]" , "B.year[14,1]" )) %>% mutate (Parameter = factor (Parameter, levels = c ("B.year[1,1]" , "B.year[2,1]" , "B.year[3,1]" , "B.year[4,1]" , "B.year[5,1]" ,"B.year[6,1]" , "B.year[7,1]" , "B.year[8,1]" , "B.year[9,1]" , "B.year[10,1]" , "B.year[11,1]" , "B.year[12,1]" , "B.year[13,1]" , "B.year[14,1]" ))) %>% ggs_caterpillar (sort = FALSE ) + scale_y_discrete (labels = rev (sort (unique (dat$ year))), limits = rev) + ylab ("Year effects, offsets to intercepts" ) + theme_bw () + geom_vline (xintercept = 0 , linetype = "dashed" )``` ##### Catchment area effects ```{r eval=FALSE} ggs_caterpillar (ggfit, family = "alpha.1" , sort = FALSE ) + scale_y_discrete (labels = rev (c ("Intercepts" , "Temp eff." , "Flow eff." , "Temp-Flow int." )), limits = rev) + ylab ("Catchment size effects" ) + theme_bw () + geom_vline (xintercept = 0 , linetype = "dashed" )``` ### Post-hoc catchment effects ```{r} <- read_csv ("data/Daymet_daily.csv" )<- climdf %>% filter (site_name %in% mysites) %>% group_by (site_name) %>% summarize (swe = mean (swe_kgm2)) %>% ungroup () %>% left_join (dat %>% group_by (site_name) %>% summarize (basin = unique (basin), area_sqmi = unique (area_sqmi), elev_ft = unique (elev_ft)) %>% ungroup ())# ggpubr::ggarrange(climdf %>% filter(site_name == "BigCreekLower") %>% ggplot(aes(x = date, y = swe_kgm2)) + geom_line(), # climdf %>% filter(site_name == "CoalCreekLower") %>% ggplot(aes(x = date, y = swe_kgm2)) + geom_line(), # climdf %>% filter(site_name == "NicolaCreek") %>% ggplot(aes(x = date, y = swe_kgm2)) + geom_line(), # climdf %>% filter(site_name == "McGeeCreekTrib") %>% ggplot(aes(x = date, y = swe_kgm2)) + geom_line(), # ncol = 1) # # climdf %>% filter(site_name == "Avery Brook", year(date) >= 2020) %>% ggplot(aes(x = date, y = daylength_sec)) + geom_line() ``` ```{r fig.width=8, fig.height=8, message=FALSE} ggpairs (basinvars %>% mutate (logarea = log (area_sqmi)) %>% left_join (dat %>% mutate (month = month (date)) %>% filter (month %in% c (8 )) %>% group_by (site_name) %>% summarize (meanSummYield = mean (Yield_mm, na.rm = TRUE ))) %>% select (basin, swe, logarea, elev_ft, meanSummYield), mapping = aes (color = basin))``` ```{r fig.width=8, fig.height=8, message=FALSE} ggpairs (basinvars %>% mutate (logarea = log (area_sqmi)) %>% left_join (dat %>% mutate (month = month (date)) %>% filter (month %in% c (8 )) %>% group_by (site_name) %>% summarize (meanSummYield = mean (Yield_mm, na.rm = TRUE ))) %>% group_by (basin) %>% mutate (z_logarea = c (scale (logarea)), z_elev_ft = c (scale (elev_ft)), z_swe = c (scale (swe)),z_yield = c (scale (meanSummYield))) %>% ungroup () %>% select (basin, z_swe, z_logarea, z_elev_ft, z_yield), mapping = aes (color = basin))``` #### PCA ##### PCA on raw variables ```{r} <- basinvars %>% mutate (logarea = log (area_sqmi)) %>% left_join (dat %>% mutate (month = month (date)) %>% filter (month %in% c (8 )) %>% group_by (site_name) %>% summarize (meanSummYield = mean (Yield_mm, na.rm = TRUE ))) %>% select (basin, site_name, swe, logarea, elev_ft, meanSummYield)<- prcomp (testdata[,- c (1 : 2 )], scale. = TRUE )``` ```{r} # scree plot fviz_eig (mypca)``` ###### PC1 ```{r} fviz_contrib (mypca, choice = "var" , axes = 1 , sort.val = "none" )``` ###### PC2 ```{r} fviz_contrib (mypca, choice = "var" , axes = 2 , sort.val = "none" )``` ###### PC3 ```{r} fviz_contrib (mypca, choice = "var" , axes = 3 , sort.val = "none" )``` ###### PC4 ```{r} fviz_contrib (mypca, choice = "var" , axes = 4 , sort.val = "none" )``` ```{r} #bi-plot fviz_pca_biplot (mypca, repel = TRUE ,col.var = "black" , # Variables color col.ind = testdata$ basin, # Individuals color addEllipses = TRUE ,#ellipse.type = "confidence", legend.title = "Basins" ``` ##### PCA on std variables ```{r} <- basinvars %>% mutate (logarea = log (area_sqmi)) %>% left_join (dat %>% mutate (month = month (date)) %>% filter (month %in% c (8 )) %>% group_by (site_name) %>% summarize (meanSummYield = mean (Yield_mm, na.rm = TRUE ))) %>% group_by (basin) %>% mutate (z_logarea = c (scale (logarea)), z_elev_ft = c (scale (elev_ft)), z_swe = c (scale (swe)),z_yield = c (scale (meanSummYield))) %>% ungroup () %>% select (basin, site_name, z_swe, z_logarea, z_elev_ft, z_yield)<- prcomp (testdata[,- c (1 : 2 )])``` ```{r} # scree plot fviz_eig (mypca_std)``` ###### PC1 ```{r} fviz_contrib (mypca_std, choice = "var" , axes = 1 , sort.val = "none" )``` ###### PC2 ```{r} fviz_contrib (mypca_std, choice = "var" , axes = 2 , sort.val = "none" )``` ###### PC3 ```{r} fviz_contrib (mypca_std, choice = "var" , axes = 3 , sort.val = "none" )``` ###### PC4 ```{r} fviz_contrib (mypca_std, choice = "var" , axes = 4 , sort.val = "none" )``` ```{r} #bi-plot fviz_pca_biplot (mypca_std, repel = TRUE ,col.var = "black" , # Variables color col.ind = testdata$ basin # Individuals color ``` #### Intercepts ```{r fig.width=9, fig.height=5} <- ggfit %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",1]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",1]" , sep = "" ))) %>% group_by (Parameter) %>% summarize (mean = mean (value)) %>% ungroup () %>% mutate (site_name = mysites) %>% left_join (basinvars) %>% left_join (dat %>% mutate (month = month (date)) %>% filter (month %in% c (8 )) %>% group_by (site_name) %>% summarize (meanSummYield = mean (Yield_mm, na.rm = TRUE ))) %>% group_by (basin) %>% mutate (z_area = scale (log (area_sqmi)), z_elev = scale (elev_ft), z_swe = scale (swe),z_yield = scale (meanSummYield)) %>% ungroup ()summary (lmer (mean ~ z_elev * z_yield + (1 + z_elev * z_yield | basin), data = mydf))ranef (lmer (mean ~ z_elev * z_yield + (1 + z_elev * z_yield | basin), data = mydf))<- mydf %>% ggplot (aes (x = log (area_sqmi), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("Catchment area (log scale)" ) + ylab ("Intercept (posterior mean)" ) #+ #ggpubr::stat_cor() <- mydf %>% ggplot (aes (x = elev_ft, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("Elevation (feet)" ) + ylab ("Intercept (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = swe, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("SWE (10-yr mean)" ) + ylab ("Intercept (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = log (meanSummYield), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("Mean August yield (log scale)" ) + ylab ("Intercept (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ()):: ggarrange (p1, p2, p3, p4, nrow = 1 , common.legend = TRUE , legend = "top" )``` ```{r fig.width=9, fig.height=9} <- mydf %>% ggplot (aes (x = log (area_sqmi), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("Catchment area (log scale)" ) + ylab ("Intercept (posterior mean)" ) #+ #ggpubr::stat_cor() <- mydf %>% ggplot (aes (x = elev_ft, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("Elevation (feet)" ) + ylab ("Intercept (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = swe, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("SWE (10-yr mean)" ) + ylab ("Intercept (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = log (meanSummYield), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("Mean August yield (log scale)" ) + ylab ("Intercept (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ()):: ggarrange (p1, p2, p3, p4, nrow = 1 )``` ##### by PC scores ```{r} <- ggfit %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",1]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",1]" , sep = "" ))) %>% group_by (Parameter) %>% summarize (mean = mean (value)) %>% ungroup () %>% mutate (site_name = mysites,PC1 = mypca_std$ x[,1 ],PC2 = mypca_std$ x[,2 ],PC3 = mypca_std$ x[,3 ],PC4 = mypca_std$ x[,4 ]) %>% left_join (basinvars %>% select (basin, site_name)) <- mydf %>% ggplot (aes (x = PC1, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + ylab ("Intercept (posterior mean)" ) #+ #ggpubr::stat_cor() <- mydf %>% ggplot (aes (x = PC2, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + ylab ("Intercept (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = PC3, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + ylab ("Intercept (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = PC3, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + ylab ("Intercept (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ()):: ggarrange (p1, p2, p3, p4, nrow = 1 , common.legend = TRUE , legend = "top" )``` #### Temp effect ```{r fig.width=9, fig.height=5} <- ggfit %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",2]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",2]" , sep = "" ))) %>% group_by (Parameter) %>% summarize (mean = mean (value)) %>% ungroup () %>% mutate (site_name = mysites) %>% left_join (basinvars) %>% left_join (dat %>% mutate (month = month (date)) %>% filter (month %in% c (8 )) %>% group_by (site_name) %>% summarize (meanSummYield = mean (Yield_mm, na.rm = TRUE ))) %>% group_by (basin) %>% mutate (z_area = scale (log (area_sqmi)), z_elev = scale (elev_ft), z_swe = scale (swe),z_yield = scale (meanSummYield)) %>% ungroup ()summary (lmer (mean ~ z_elev * z_yield + (1 + z_elev * z_yield | basin), data = mydf))ranef (lmer (mean ~ z_elev * z_yield + (1 + z_elev * z_yield | basin), data = mydf))<- mydf %>% ggplot (aes (x = log (area_sqmi), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("Catchment area (log scale)" ) + ylab ("Temp effect (posterior mean)" ) #+ ggpubr::stat_cor() <- mydf %>% ggplot (aes (x = elev_ft, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("Elevation (feet)" ) + ylab ("Temp effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = swe, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("SWE (10-yr mean)" ) + ylab ("Temp effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = log (meanSummYield), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("Mean August yield (log scale)" ) + ylab ("Temp effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ()):: ggarrange (p1, p2, p3, p4, nrow = 1 , common.legend = TRUE , legend = "top" )``` ```{r fig.width=9, fig.height=9} <- mydf %>% ggplot (aes (x = log (area_sqmi), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("Catchment area (log scale)" ) + ylab ("Temp effect (posterior mean)" ) #+ ggpubr::stat_cor() <- mydf %>% ggplot (aes (x = elev_ft, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("Elevation (feet)" ) + ylab ("Temp effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = swe, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("SWE (10-yr mean)" ) + ylab ("Temp effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = log (meanSummYield), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("Mean August yield (log scale)" ) + ylab ("Temp effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ()):: ggarrange (p1, p2, p3, p4, nrow = 1 )``` ##### by PC scores ```{r} <- ggfit %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",2]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",2]" , sep = "" ))) %>% group_by (Parameter) %>% summarize (mean = mean (value)) %>% ungroup () %>% mutate (site_name = mysites,PC1 = mypca_std$ x[,1 ],PC2 = mypca_std$ x[,2 ],PC3 = mypca_std$ x[,3 ],PC4 = mypca_std$ x[,4 ]) %>% left_join (basinvars %>% select (basin, site_name)) <- mydf %>% ggplot (aes (x = PC1, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + ylab ("Temp effect (posterior mean)" ) #+ #ggpubr::stat_cor() <- mydf %>% ggplot (aes (x = PC2, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + ylab ("Temp effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = PC3, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + ylab ("Temp effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = PC3, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + ylab ("Temp effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ()):: ggarrange (p1, p2, p3, p4, nrow = 1 , common.legend = TRUE , legend = "top" )``` #### Flow effect ```{r fig.width=9, fig.height=5} <- ggfit %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",3]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",3]" , sep = "" ))) %>% group_by (Parameter) %>% summarize (mean = mean (value)) %>% ungroup () %>% mutate (site_name = mysites) %>% left_join (basinvars) %>% left_join (dat %>% mutate (month = month (date)) %>% filter (month %in% c (8 )) %>% group_by (site_name) %>% summarize (meanSummYield = mean (Yield_mm, na.rm = TRUE ))) %>% group_by (basin) %>% mutate (z_area = scale (log (area_sqmi)), z_elev = scale (elev_ft), z_swe = scale (swe),z_yield = scale (meanSummYield)) %>% ungroup ()summary (lmer (mean ~ z_elev * z_yield + (1 + z_elev * z_yield | basin), data = mydf))ranef (lmer (mean ~ z_elev * z_yield + (1 + z_elev * z_yield | basin), data = mydf))<- mydf %>% ggplot (aes (x = log (area_sqmi), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("Catchment area (log scale)" ) + ylab ("Flow effect (posterior mean)" ) #+ ggpubr::stat_cor() <- mydf %>% ggplot (aes (x = elev_ft, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("Elevation (feet)" ) + ylab ("Flow effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = swe, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("SWE (10-yr mean)" ) + ylab ("Flow effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = log (meanSummYield), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("Mean August yield (log scale)" ) + ylab ("Flow effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ()):: ggarrange (p1, p2, p3, p4, nrow = 1 , common.legend = TRUE , legend = "top" )``` ```{r fig.width=9, fig.height=9} <- mydf %>% ggplot (aes (x = log (area_sqmi), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("Catchment area (log scale)" ) + ylab ("Flow effect (posterior mean)" ) #+ ggpubr::stat_cor() <- mydf %>% ggplot (aes (x = elev_ft, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("Elevation (feet)" ) + ylab ("Flow effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = swe, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("SWE (10-yr mean)" ) + ylab ("Flow effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = log (meanSummYield), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("Mean August yield (log scale)" ) + ylab ("Flow effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ()):: ggarrange (p1, p2, p3, p4, nrow = 1 )``` ##### by PC scores ```{r} <- ggfit %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",3]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",3]" , sep = "" ))) %>% group_by (Parameter) %>% summarize (mean = mean (value)) %>% ungroup () %>% mutate (site_name = mysites,PC1 = mypca_std$ x[,1 ],PC2 = mypca_std$ x[,2 ],PC3 = mypca_std$ x[,3 ],PC4 = mypca_std$ x[,4 ]) %>% left_join (basinvars %>% select (basin, site_name)) <- mydf %>% ggplot (aes (x = PC1, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + ylab ("Flow effect (posterior mean)" )<- mydf %>% ggplot (aes (x = PC2, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + ylab ("Flow effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = PC3, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + ylab ("Flow effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = PC3, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + ylab ("Flow effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ()):: ggarrange (p1, p2, p3, p4, nrow = 1 , common.legend = TRUE , legend = "top" )``` #### Air temp lagged 1 ```{r fig.width=9, fig.height=5} <- ggfit %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",5]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",5]" , sep = "" ))) %>% group_by (Parameter) %>% summarize (mean = mean (value)) %>% ungroup () %>% mutate (site_name = mysites) %>% left_join (basinvars) %>% left_join (dat %>% mutate (month = month (date)) %>% filter (month %in% c (8 )) %>% group_by (site_name) %>% summarize (meanSummYield = mean (Yield_mm, na.rm = TRUE ))) %>% group_by (basin) %>% mutate (z_area = scale (log (area_sqmi)), z_elev = scale (elev_ft), z_swe = scale (swe),z_yield = scale (meanSummYield)) %>% ungroup ()summary (lmer (mean ~ z_elev * z_yield + (1 + z_elev * z_yield | basin), data = mydf))ranef (lmer (mean ~ z_elev * z_yield + (1 + z_elev * z_yield | basin), data = mydf))<- mydf %>% ggplot (aes (x = log (area_sqmi), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("Catchment area (log scale)" ) + ylab ("AirT lag-1 effect (posterior mean)" ) #+ ggpubr::stat_cor() <- mydf %>% ggplot (aes (x = elev_ft, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("Elevation (feet)" ) + ylab ("AirT lag-1 effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = swe, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("SWE (10-yr mean)" ) + ylab ("AirT lag-1 effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = log (meanSummYield), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("Mean August yield (log scale)" ) + ylab ("AirT lag-1 effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ()):: ggarrange (p1, p2, p3, p4, nrow = 1 , common.legend = TRUE , legend = "top" )``` ```{r fig.width=9, fig.height=9} <- mydf %>% ggplot (aes (x = log (area_sqmi), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("Catchment area (log scale)" ) + ylab ("AirT lag-1 effect (posterior mean)" ) #+ ggpubr::stat_cor() <- mydf %>% ggplot (aes (x = elev_ft, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("Elevation (feet)" ) + ylab ("AirT lag-1 effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = swe, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("SWE (10-yr mean)" ) + ylab ("AirT lag-1 effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = log (meanSummYield), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("Mean August yield (log scale)" ) + ylab ("AirT lag-1 effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ()):: ggarrange (p1, p2, p3, p4, nrow = 1 )``` #### Air temp lagged 2 ```{r fig.width=9, fig.height=5} <- ggfit %>% filter (Parameter %in% paste ("B.site[" , c (1 : jags.data$ nsites), ",6]" , sep = "" )) %>% mutate (Parameter = factor (Parameter, levels = paste ("B.site[" , c (1 : jags.data$ nsites), ",6]" , sep = "" ))) %>% group_by (Parameter) %>% summarize (mean = mean (value)) %>% ungroup () %>% mutate (site_name = mysites) %>% left_join (basinvars) %>% left_join (dat %>% mutate (month = month (date)) %>% filter (month %in% c (8 )) %>% group_by (site_name) %>% summarize (meanSummYield = mean (Yield_mm, na.rm = TRUE ))) %>% group_by (basin) %>% mutate (z_area = scale (log (area_sqmi)), z_elev = scale (elev_ft), z_swe = scale (swe),z_yield = scale (meanSummYield)) %>% ungroup ()summary (lmer (mean ~ z_elev * z_yield + (1 + z_elev * z_yield | basin), data = mydf))ranef (lmer (mean ~ z_elev * z_yield + (1 + z_elev * z_yield | basin), data = mydf))<- mydf %>% ggplot (aes (x = log (area_sqmi), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("Catchment area (log scale)" ) + ylab ("AirT lag-2 effect (posterior mean)" ) #+ ggpubr::stat_cor() <- mydf %>% ggplot (aes (x = elev_ft, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("Elevation (feet)" ) + ylab ("AirT lag-2 effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = swe, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("SWE (10-yr mean)" ) + ylab ("AirT lag-2 effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = log (meanSummYield), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + xlab ("Mean August yield (log scale)" ) + ylab ("AirT lag-2 effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ()):: ggarrange (p1, p2, p3, p4, nrow = 1 , common.legend = TRUE , legend = "top" )``` ```{r fig.width=9, fig.height=9} <- mydf %>% ggplot (aes (x = log (area_sqmi), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("Catchment area (log scale)" ) + ylab ("AirT lag-2 effect (posterior mean)" ) #+ ggpubr::stat_cor() <- mydf %>% ggplot (aes (x = elev_ft, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("Elevation (feet)" ) + ylab ("AirT lag-2 effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = swe, y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("SWE (10-yr mean)" ) + ylab ("AirT lag-2 effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ())<- mydf %>% ggplot (aes (x = log (meanSummYield), y = mean, color = basin)) + geom_smooth (method = "lm" ) + geom_point () + theme_bw () + theme (legend.position = "none" ) + facet_wrap (~ basin, scales = "free" , ncol = 1 ) + xlab ("Mean August yield (log scale)" ) + ylab ("AirT lag-2 effect (posterior mean)" ) + #ggpubr::stat_cor() + theme (axis.title.y = element_blank (), axis.text.y = element_blank ()):: ggarrange (p1, p2, p3, p4, nrow = 1 )```