Code
rcdat <- read_csv("Data/Derived/ReddCounts_WGFD_1971-2021_cleaned_withFlowTempCovariates_SeparateFlowComponents_age0.csv") %>% filter(!is.na(reddsperkm)) %>% select(year)
rcsum <- read_csv("ReddCounts_DataSummary.csv")Purpose: Explore covariate time series data data - trends and step-changes in environmental variables describing climate and dam management.
Redd count data to get years for rug marks
rcdat <- read_csv("Data/Derived/ReddCounts_WGFD_1971-2021_cleaned_withFlowTempCovariates_SeparateFlowComponents_age0.csv") %>% filter(!is.na(reddsperkm)) %>% select(year)
rcsum <- read_csv("ReddCounts_DataSummary.csv")Covariate data
jldramp <- read_csv("Data/Derived/JLD_RampDown_Summary_CalendarYear_1960-2022.csv") #%>% rename(broodyr = year)
jldflow <- read_csv("Data/Derived/JLD_ManagedFlow_Covariates_BroodYear_1960-2022.csv")
natflow <- read_csv("Data/Derived/SnakeTribs_NaturalFlow_Covariates_BroodYear_1960-2022.csv") %>% rename(flowstn = site) %>% filter(flowstn %in% c("SnakeBeFlat", "SnakeNat"))
airtemp <- read_csv("Data/Derived/AirTemperature_Covariates_BroodYear_1960-2022.csv") %>% filter(site %in% c("moose")) %>% rename(tempstn = site)Misc.
# color palette
mycols <- c("#E69F00", "#56B4E9", "#009E73", "#999999")
# covariate names
names.covars.new <- c("Duration of ramp down",
"Timing of ramp down",
"Mgd. winter flow variation",
"Mgd. summer mean flow",
"Mgd. peak flow mag. (0)",
"Mgd. peak flow mag. (1)",
"Mgd. peak flow timing",
"Nat. peak flow magnitude",
"Nat. peak flow timing",
"Autumn temperature",
"Winter temperature",
"Spring temperature",
"Summer temperature",
"Mgd. x Nat. peak flow timing")plotlist <- list()Test for change before/after 1989
pre <- unlist(jldramp %>% filter(broodyr < 1989) %>% select(jld_rampdur))
post <- unlist(jldramp %>% filter(broodyr >= 1989) %>% select(jld_rampdur))
prepost <- tibble(period = factor(c(rep("pre", times = length(pre)), rep("post", times = length(post))), levels = c("pre", "post")),
value = c(unlist(pre), unlist(post)))
t.test(pre, post, alternative = "two.sided")
Welch Two Sample t-test
data: pre and post
t = 1.3686, df = 36.044, p-value = 0.1796
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-0.9841648 5.0693575
sample estimates:
mean of x mean of y
10.689655 8.647059 var.test(pre, post, alternative = "two.sided")
F test to compare two variances
data: pre and post
F = 5.898, num df = 28, denom df = 33, p-value = 2.834e-06
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
2.885426 12.321171
sample estimates:
ratio of variances
5.89802 Create plot
plotlist[[1]] <- eval(substitute(ggplot() +
geom_point(data = jldramp, mapping = aes(x = broodyr, y = jld_rampdur), shape = 16) +
geom_line(data = jldramp, mapping = aes(x = broodyr, y = jld_rampdur)) +
geom_ysideboxplot(data = prepost, mapping = aes(x = period, y = value), orientation = "x", fill = "grey80") +
xlab("Year") + ylab("Duration of ramp down (days)") +
geom_rug(data = rcdat, mapping = aes(x = jitter(year, factor = 1.5))) +
geom_vline(xintercept = 1989, color = "red", linetype = "dashed", linewidth = 0.7) +
geom_vline(xintercept = 1996, color = "grey50", linetype = "dashed") +
theme_bw() +
theme(panel.grid = element_blank(), axis.text = element_text(color = "black"),
panel.border = element_rect(color = mycols[1], fill = NA, linewidth = 2),
ggside.panel.border = element_blank(), ggside.axis.text = element_blank(),
ggside.axis.ticks = element_blank(), axis.text.y = element_text(angle = 90, hjust = 0.5, vjust = 0.5))))Test for change before/after 1989
pre <- unlist(jldramp %>% filter(broodyr < 1989) %>% select(jld_rampratemindoy))
post <- unlist(jldramp %>% filter(broodyr >= 1989) %>% select(jld_rampratemindoy))
prepost <- tibble(period = factor(c(rep("pre", times = length(pre)), rep("post", times = length(post))), levels = c("pre", "post")),
value = c(unlist(pre), unlist(post)))
t.test(pre, post, alternative = "two.sided")
Welch Two Sample t-test
data: pre and post
t = -5.1168, df = 31.432, p-value = 1.482e-05
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-13.622019 -5.860739
sample estimates:
mean of x mean of y
264.7586 274.5000 var.test(pre, post, alternative = "two.sided")
F test to compare two variances
data: pre and post
F = 13.938, num df = 28, denom df = 33, p-value = 3.494e-11
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
6.818913 29.117712
sample estimates:
ratio of variances
13.93836 Create plot
plotlist[[2]] <- eval(substitute(ggplot() +
geom_point(data = jldramp, mapping = aes(x = broodyr, y = jld_rampratemindoy), shape = 16) +
geom_line(data = jldramp, mapping = aes(x = broodyr, y = jld_rampratemindoy)) +
geom_ysideboxplot(data = prepost, mapping = aes(x = period, y = value), orientation = "x", fill = "grey80") +
xlab("Year") + ylab("Timing of ramp down") +
geom_rug(data = rcdat, mapping = aes(x = jitter(year, factor = 1.5))) +
geom_vline(xintercept = 1989, color = "red", linetype = "dashed", linewidth = 0.7) +
geom_vline(xintercept = 1996, color = "grey50", linetype = "dashed") +
scale_y_continuous(breaks = c(245, 254, 264, 274, 284), labels = c("1 Sept", "10 Sept", "20 Sept", "30 Sept", "10 Oct")) +
theme_bw() +
theme(panel.grid = element_blank(), axis.text = element_text(color = "black"),
panel.border = element_rect(color = mycols[1], fill = NA, linewidth = 2),
ggside.panel.border = element_blank(), ggside.axis.text = element_blank(),
ggside.axis.ticks = element_blank(), axis.text.y = element_text(angle = 90, hjust = 0.5, vjust = 0.5))))Test for change before/after 1989
pre <- unlist(jldflow %>% filter(broodyr < 1989) %>% select(jld_winvar))
post <- unlist(jldflow %>% filter(broodyr >= 1989) %>% select(jld_winvar))
prepost <- tibble(period = factor(c(rep("pre", times = length(pre)), rep("post", times = length(post))), levels = c("pre", "post")),
value = c(unlist(pre), unlist(post)))
t.test(pre, post, alternative = "two.sided")
Welch Two Sample t-test
data: pre and post
t = 4.8455, df = 29.257, p-value = 3.818e-05
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
0.04390844 0.10800512
sample estimates:
mean of x mean of y
0.09192974 0.01597296 var.test(pre, post, alternative = "two.sided")
F test to compare two variances
data: pre and post
F = 38.041, num df = 28, denom df = 33, p-value < 2.2e-16
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
18.61031 79.46862
sample estimates:
ratio of variances
38.04083 Create plot
plotlist[[3]] <- eval(substitute(ggplot() +
geom_point(data = jldflow, mapping = aes(x = broodyr, y = jld_winvar), shape = 16) +
geom_line(data = jldflow, mapping = aes(x = broodyr, y = jld_winvar)) +
geom_ysideboxplot(data = prepost, mapping = aes(x = period, y = value), orientation = "x", fill = "grey80") +
xlab("Year") + ylab("Mgd. winter flow variation") +
geom_rug(data = rcdat, mapping = aes(x = jitter(year, factor = 1.5))) +
geom_vline(xintercept = 1989, color = "red", linetype = "dashed", linewidth = 0.7) +
geom_vline(xintercept = 1996, color = "grey50", linetype = "dashed") +
theme_bw() +
theme(panel.grid = element_blank(), axis.text = element_text(color = "black"),
panel.border = element_rect(color = mycols[1], fill = NA, linewidth = 2),
ggside.panel.border = element_blank(), ggside.axis.text = element_blank(),
ggside.axis.ticks = element_blank(), axis.text.y = element_text(angle = 90, hjust = 0.5, vjust = 0.5))))Test for change before/after 1989
pre <- unlist(jldflow %>% filter(broodyr < 1989) %>% select(jld_summean))
post <- unlist(jldflow %>% filter(broodyr >= 1989) %>% select(jld_summean))
prepost <- tibble(period = factor(c(rep("pre", times = length(pre)), rep("post", times = length(post))), levels = c("pre", "post")),
value = c(unlist(pre), unlist(post)))
t.test(pre, post, alternative = "two.sided")
Welch Two Sample t-test
data: pre and post
t = 0.62431, df = 60.481, p-value = 0.5348
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-289.0549 551.4150
sample estimates:
mean of x mean of y
2840.345 2709.165 var.test(pre, post, alternative = "two.sided")
F test to compare two variances
data: pre and post
F = 0.87075, num df = 28, denom df = 33, p-value = 0.7137
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
0.4259891 1.8190332
sample estimates:
ratio of variances
0.8707529 Create plot
plotlist[[4]] <- eval(substitute(ggplot() +
geom_point(data = jldflow, mapping = aes(x = broodyr, y = jld_summean), shape = 16) +
geom_line(data = jldflow, mapping = aes(x = broodyr, y = jld_summean)) +
geom_ysideboxplot(data = prepost, mapping = aes(x = period, y = value), orientation = "x", fill = "grey80") +
xlab("Year") + ylab("Mgd. summer mean flow (cfs)") +
geom_rug(data = rcdat, mapping = aes(x = jitter(year, factor = 1.5))) +
geom_vline(xintercept = 1989, color = "red", linetype = "dashed", linewidth = 0.7) +
geom_vline(xintercept = 1996, color = "grey50", linetype = "dashed") +
theme_bw() +
theme(panel.grid = element_blank(), axis.text = element_text(color = "black"),
panel.border = element_rect(color = mycols[1], fill = NA, linewidth = 2),
ggside.panel.border = element_blank(), ggside.axis.text = element_blank(),
ggside.axis.ticks = element_blank(), axis.text.y = element_text(angle = 90, hjust = 0.5, vjust = 0.5))))Test for change before/after 1989
pre <- unlist(jldflow %>% filter(broodyr < 1989) %>% select(jld_peakmag))
post <- unlist(jldflow %>% filter(broodyr >= 1989) %>% select(jld_peakmag))
prepost <- tibble(period = factor(c(rep("pre", times = length(pre)), rep("post", times = length(post))), levels = c("pre", "post")),
value = c(unlist(pre), unlist(post)))
t.test(pre, post, alternative = "two.sided")
Welch Two Sample t-test
data: pre and post
t = 0.58672, df = 60.976, p-value = 0.5596
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-658.1472 1204.7395
sample estimates:
mean of x mean of y
5972.414 5699.118 var.test(pre, post, alternative = "two.sided")
F test to compare two variances
data: pre and post
F = 0.69515, num df = 28, denom df = 33, p-value = 0.3295
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
0.3400795 1.4521868
sample estimates:
ratio of variances
0.6951472 Create plot
plotlist[[5]] <- eval(substitute(ggplot() +
geom_point(data = jldflow, mapping = aes(x = broodyr, y = jld_peakmag), shape = 16) +
geom_line(data = jldflow, mapping = aes(x = broodyr, y = jld_peakmag)) +
geom_ysideboxplot(data = prepost, mapping = aes(x = period, y = value), orientation = "x", fill = "grey80") +
xlab("Year") + ylab("Mgd. peak flow magnitude (cfs)") +
geom_rug(data = rcdat, mapping = aes(x = jitter(year, factor = 1.5))) +
geom_vline(xintercept = 1989, color = "red", linetype = "dashed", linewidth = 0.7) +
geom_vline(xintercept = 1996, color = "grey50", linetype = "dashed") +
theme_bw() +
theme(panel.grid = element_blank(), axis.text = element_text(color = "black"),
panel.border = element_rect(color = mycols[1], fill = NA, linewidth = 2),
ggside.panel.border = element_blank(), ggside.axis.text = element_blank(),
ggside.axis.ticks = element_blank(), axis.text.y = element_text(angle = 90, hjust = 0.5, vjust = 0.5))))Test for change before/after 1989
pre <- unlist(jldflow %>% filter(broodyr < 1989) %>% select(jld_peaktime))
post <- unlist(jldflow %>% filter(broodyr >= 1989) %>% select(jld_peaktime))
prepost <- tibble(period = factor(c(rep("pre", times = length(pre)), rep("post", times = length(post))), levels = c("pre", "post")),
value = c(unlist(pre), unlist(post)))
t.test(pre, post, alternative = "two.sided")
Welch Two Sample t-test
data: pre and post
t = 1.6982, df = 45.781, p-value = 0.09626
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-1.34816 15.88446
sample estimates:
mean of x mean of y
291.1481 283.8800 var.test(pre, post, alternative = "two.sided")
F test to compare two variances
data: pre and post
F = 2.2094, num df = 26, denom df = 24, p-value = 0.05475
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
0.9835477 4.8992022
sample estimates:
ratio of variances
2.209417 Create plot
plotlist[[6]] <- eval(substitute(ggplot() +
geom_point(data = jldflow, mapping = aes(x = broodyr, y = jld_peaktime), shape = 16) +
geom_line(data = jldflow, mapping = aes(x = broodyr, y = jld_peaktime)) +
geom_ysideboxplot(data = prepost, mapping = aes(x = period, y = value), orientation = "x", fill = "grey80") +
xlab("Year") + ylab("Mgd. peak flow timing") +
geom_rug(data = rcdat, mapping = aes(x = jitter(year, factor = 1.5))) +
geom_vline(xintercept = 1989, color = "red", linetype = "dashed", linewidth = 0.7) +
geom_vline(xintercept = 1996, color = "grey50", linetype = "dashed") +
scale_y_continuous(breaks = c(242, 273, 303), labels = c("1 May", "1 June", "1 July")) +
theme_bw() +
theme(panel.grid = element_blank(), axis.text = element_text(color = "black"),
panel.border = element_rect(color = mycols[1], fill = NA, linewidth = 2),
ggside.panel.border = element_blank(), ggside.axis.text = element_blank(),
ggside.axis.ticks = element_blank(), axis.text.y = element_text(angle = 90, hjust = 0.5, vjust = 0.5))))Test for linear trend
mod <- lm(natq_peakmag ~ broodyr, natflow %>% filter(flowstn == "SnakeNat"))
summary(mod)
Call:
lm(formula = natq_peakmag ~ broodyr, data = natflow %>% filter(flowstn ==
"SnakeNat"))
Residuals:
Min 1Q Median 3Q Max
-6702.8 -2599.9 -538.2 1332.4 8620.0
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 36446.36 75748.89 0.481 0.633
broodyr -12.76 37.90 -0.337 0.738
Residual standard error: 3638 on 46 degrees of freedom
Multiple R-squared: 0.002457, Adjusted R-squared: -0.01923
F-statistic: 0.1133 on 1 and 46 DF, p-value: 0.7379Create plot
plotlist[[7]] <- eval(substitute(ggplot() +
geom_smooth(data = natflow %>% filter(flowstn == "SnakeNat"), mapping = aes(x = broodyr, y = natq_peakmag), color = mycols[2]) +
geom_point(data = natflow %>% filter(flowstn == "SnakeNat"), mapping = aes(x = broodyr, y = natq_peakmag), shape = 16) +
geom_line(data = natflow %>% filter(flowstn == "SnakeNat"), mapping = aes(x = broodyr, y = natq_peakmag)) +
xlab("Year") + ylab("Nat. peak flow magnitude (cfs)") +
geom_rug(data = rcdat, mapping = aes(x = jitter(year, factor = 1.5))) +
xlim(1960,2022) +
theme_bw() +
theme(panel.grid = element_blank(), axis.text = element_text(color = "black"),
panel.border = element_rect(color = mycols[2], fill = NA, linewidth = 2),
ggside.panel.border = element_blank(), ggside.axis.text = element_blank(),
ggside.axis.ticks = element_blank(), axis.text.y = element_text(angle = 90, hjust = 0.5, vjust = 0.5))))Test for linear trend
mod <- lm(natq_peaktime ~ broodyr, natflow %>% filter(flowstn == "SnakeNat"))
summary(mod)
Call:
lm(formula = natq_peaktime ~ broodyr, data = natflow %>% filter(flowstn ==
"SnakeNat"))
Residuals:
Min 1Q Median 3Q Max
-25.945 -5.880 2.588 6.668 26.866
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 362.88450 249.39101 1.455 0.152
broodyr -0.04266 0.12479 -0.342 0.734
Residual standard error: 11.98 on 46 degrees of freedom
Multiple R-squared: 0.002534, Adjusted R-squared: -0.01915
F-statistic: 0.1169 on 1 and 46 DF, p-value: 0.734Create plot
plotlist[[8]] <- eval(substitute(ggplot() +
geom_smooth(data = natflow %>% filter(flowstn == "SnakeNat"), mapping = aes(x = broodyr, y = natq_peaktime), color = mycols[2]) +
geom_point(data = natflow %>% filter(flowstn == "SnakeNat"), mapping = aes(x = broodyr, y = natq_peaktime), shape = 16) +
geom_line(data = natflow %>% filter(flowstn == "SnakeNat"), mapping = aes(x = broodyr, y = natq_peaktime)) +
xlab("Year") + ylab("Nat. peak flow timing") +
geom_rug(data = rcdat, mapping = aes(x = jitter(year, factor = 1.5))) +
scale_y_continuous(breaks = c(256, 273, 287, 303), labels = c("15 May", "1 June", "15 June", "1 July")) +
xlim(1960,2022) +
theme_bw() +
theme(panel.grid = element_blank(), axis.text = element_text(color = "black"),
panel.border = element_rect(color = mycols[2], fill = NA, linewidth = 2),
ggside.panel.border = element_blank(), ggside.axis.text = element_blank(),
ggside.axis.ticks = element_blank(), axis.text.y = element_text(angle = 90, hjust = 0.5, vjust = 0.5))))Test for linear trend, entire time series
summary(lm(temp_falmean ~ broodyr, airtemp))
Call:
lm(formula = temp_falmean ~ broodyr, data = airtemp)
Residuals:
Min 1Q Median 3Q Max
-2.91124 -0.70124 0.05212 0.54642 2.65488
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -24.557614 15.257840 -1.61 0.1128
broodyr 0.014405 0.007663 1.88 0.0651 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.1 on 59 degrees of freedom
(2 observations deleted due to missingness)
Multiple R-squared: 0.05651, Adjusted R-squared: 0.04052
F-statistic: 3.534 on 1 and 59 DF, p-value: 0.06506Since 1980
summary(lm(temp_falmean ~ broodyr, airtemp %>% filter(broodyr >= 1980)))
Call:
lm(formula = temp_falmean ~ broodyr, data = airtemp %>% filter(broodyr >=
1980))
Residuals:
Min 1Q Median 3Q Max
-2.81524 -0.38704 -0.09419 0.62226 1.74131
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -93.98092 22.11287 -4.250 0.000124 ***
broodyr 0.04903 0.01105 4.437 6.97e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.8992 on 40 degrees of freedom
(1 observation deleted due to missingness)
Multiple R-squared: 0.3299, Adjusted R-squared: 0.3131
F-statistic: 19.69 on 1 and 40 DF, p-value: 6.971e-05Create plot
plotlist[[9]] <- eval(substitute(ggplot() +
geom_smooth(data = airtemp, mapping = aes(x = broodyr, y = temp_falmean), color = mycols[3]) +
geom_point(data = airtemp, mapping = aes(x = broodyr, y = temp_falmean), shape = 16) +
geom_line(data = airtemp, mapping = aes(x = broodyr, y = temp_falmean)) +
xlab("Year") + ylab(expression(paste("Autumn temperature ("^"o", "C)", sep = ""))) +
geom_rug(data = rcdat, mapping = aes(x = jitter(year, factor = 1.5))) +
xlim(1960,2022) +
theme_bw() +
theme(panel.grid = element_blank(), axis.text = element_text(color = "black"),
panel.border = element_rect(color = mycols[3], fill = NA, linewidth = 2),
ggside.panel.border = element_blank(), ggside.axis.text = element_blank(),
ggside.axis.ticks = element_blank(), axis.text.y = element_text(angle = 90, hjust = 0.5, vjust = 0.5))))Test for linear trend, entire time series
summary(lm(temp_winmean ~ broodyr, airtemp))
Call:
lm(formula = temp_winmean ~ broodyr, data = airtemp)
Residuals:
Min 1Q Median 3Q Max
-4.2967 -0.9897 0.2001 0.9525 3.7107
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -46.67592 23.98767 -1.946 0.0564 .
broodyr 0.01879 0.01205 1.559 0.1241
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.736 on 60 degrees of freedom
(1 observation deleted due to missingness)
Multiple R-squared: 0.03895, Adjusted R-squared: 0.02294
F-statistic: 2.432 on 1 and 60 DF, p-value: 0.1241Since 1980
summary(lm(temp_winmean ~ broodyr, airtemp %>% filter(broodyr >= 1980)))
Call:
lm(formula = temp_winmean ~ broodyr, data = airtemp %>% filter(broodyr >=
1980))
Residuals:
Min 1Q Median 3Q Max
-3.3223 -1.2765 0.0218 0.8584 4.8067
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -133.25545 41.20282 -3.234 0.00245 **
broodyr 0.06196 0.02059 3.009 0.00452 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.675 on 40 degrees of freedom
(1 observation deleted due to missingness)
Multiple R-squared: 0.1846, Adjusted R-squared: 0.1642
F-statistic: 9.056 on 1 and 40 DF, p-value: 0.004517Create plot
plotlist[[10]] <- eval(substitute(ggplot() +
geom_smooth(data = airtemp, mapping = aes(x = broodyr, y = temp_winmean), color = mycols[3]) +
geom_point(data = airtemp, mapping = aes(x = broodyr, y = temp_winmean), shape = 16) +
geom_line(data = airtemp, mapping = aes(x = broodyr, y = temp_winmean)) +
xlab("Year") + ylab(expression(paste("Winter temperature ("^"o", "C)", sep = ""))) +
geom_rug(data = rcdat, mapping = aes(x = jitter(year, factor = 1.5))) +
xlim(1960,2022) +
theme_bw() +
theme(panel.grid = element_blank(), axis.text = element_text(color = "black"),
panel.border = element_rect(color = mycols[3], fill = NA, linewidth = 2),
ggside.panel.border = element_blank(), ggside.axis.text = element_blank(),
ggside.axis.ticks = element_blank(), axis.text.y = element_text(angle = 90, hjust = 0.5, vjust = 0.5))))Test for linear trend, entire time series
summary(lm(temp_sprmean ~ broodyr, airtemp))
Call:
lm(formula = temp_sprmean ~ broodyr, data = airtemp)
Residuals:
Min 1Q Median 3Q Max
-2.8511 -0.9543 -0.1377 0.8293 3.0930
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -44.305784 16.790930 -2.639 0.01071 *
broodyr 0.023415 0.008432 2.777 0.00741 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.204 on 57 degrees of freedom
(4 observations deleted due to missingness)
Multiple R-squared: 0.1192, Adjusted R-squared: 0.1037
F-statistic: 7.712 on 1 and 57 DF, p-value: 0.007413Since 1980
summary(lm(temp_sprmean ~ broodyr, airtemp %>% filter(broodyr >= 1980)))
Call:
lm(formula = temp_sprmean ~ broodyr, data = airtemp %>% filter(broodyr >=
1980))
Residuals:
Min 1Q Median 3Q Max
-2.8763 -0.9135 -0.1178 0.8993 2.8489
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -16.839474 32.367288 -0.520 0.606
broodyr 0.009742 0.016168 0.603 0.550
Residual standard error: 1.246 on 38 degrees of freedom
(3 observations deleted due to missingness)
Multiple R-squared: 0.009464, Adjusted R-squared: -0.0166
F-statistic: 0.3631 on 1 and 38 DF, p-value: 0.5504Create plot
plotlist[[11]] <- eval(substitute(ggplot() +
geom_smooth(data = airtemp, mapping = aes(x = broodyr, y = temp_sprmean), color = mycols[3]) +
geom_point(data = airtemp, mapping = aes(x = broodyr, y = temp_sprmean), shape = 16) +
geom_line(data = airtemp, mapping = aes(x = broodyr, y = temp_sprmean)) +
xlab("Year") + ylab(expression(paste("Spring temperature ("^"o", "C)", sep = ""))) +
geom_rug(data = rcdat, mapping = aes(x = jitter(year, factor = 1.5))) +
xlim(1960,2022) +
theme_bw() +
theme(panel.grid = element_blank(), axis.text = element_text(color = "black"),
panel.border = element_rect(color = mycols[3], fill = NA, linewidth = 2),
ggside.panel.border = element_blank(), ggside.axis.text = element_blank(),
ggside.axis.ticks = element_blank(), axis.text.y = element_text(angle = 90, hjust = 0.5, vjust = 0.5))))Test for linear trend, entire time series
summary(lm(temp_summean ~ broodyr, airtemp))
Call:
lm(formula = temp_summean ~ broodyr, data = airtemp)
Residuals:
Min 1Q Median 3Q Max
-3.4453 -0.7008 -0.0323 0.5342 3.2195
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -38.777282 14.590886 -2.658 0.010192 *
broodyr 0.026904 0.007328 3.672 0.000533 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.029 on 57 degrees of freedom
(4 observations deleted due to missingness)
Multiple R-squared: 0.1913, Adjusted R-squared: 0.1771
F-statistic: 13.48 on 1 and 57 DF, p-value: 0.0005329Since 1980
summary(lm(temp_summean ~ broodyr, airtemp %>% filter(broodyr >= 1980)))
Call:
lm(formula = temp_summean ~ broodyr, data = airtemp %>% filter(broodyr >=
1980))
Residuals:
Min 1Q Median 3Q Max
-3.1859 -0.6489 -0.0958 0.7208 2.1869
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -83.14643 24.95748 -3.332 0.001898 **
broodyr 0.04905 0.01247 3.932 0.000335 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.011 on 39 degrees of freedom
(2 observations deleted due to missingness)
Multiple R-squared: 0.2839, Adjusted R-squared: 0.2655
F-statistic: 15.46 on 1 and 39 DF, p-value: 0.0003353Create plot
plotlist[[12]] <- eval(substitute(ggplot() +
geom_smooth(data = airtemp, mapping = aes(x = broodyr, y = temp_summean), color = mycols[3]) +
geom_point(data = airtemp, mapping = aes(x = broodyr, y = temp_summean), shape = 16) +
geom_line(data = airtemp, mapping = aes(x = broodyr, y = temp_summean)) +
xlab("Year") + ylab(expression(paste("Summer temperature ("^"o", "C)", sep = ""))) +
geom_rug(data = rcdat, mapping = aes(x = jitter(year, factor = 1.5))) +
xlim(1960,2022) +
theme_bw() +
theme(panel.grid = element_blank(), axis.text = element_text(color = "black"),
panel.border = element_rect(color = mycols[3], fill = NA, linewidth = 2),
ggside.panel.border = element_blank(), ggside.axis.text = element_blank(),
ggside.axis.ticks = element_blank(), axis.text.y = element_text(angle = 90, hjust = 0.5, vjust = 0.5))))Combine marginal effects plots for covariates with strong and weak effects only
cp <- egg::ggarrange(plotlist[[1]] + annotation_custom(grid::textGrob("(a)", 0.08, 0.92)) + theme(axis.title.x = element_blank()),
plotlist[[2]] + annotation_custom(grid::textGrob("(b)", 0.07, 0.92)) + theme(axis.title.x = element_blank()),
plotlist[[3]] + annotation_custom(grid::textGrob("(c)", 0.08, 0.92)) + theme(axis.title.x = element_blank()),
plotlist[[4]] + annotation_custom(grid::textGrob("(d)", 0.11, 0.92)) + theme(axis.title.x = element_blank()),
plotlist[[5]] + annotation_custom(grid::textGrob("(e)", 0.08, 0.92)) + theme(axis.title.x = element_blank()),
plotlist[[6]] + annotation_custom(grid::textGrob("(f)", 0.06, 0.92)) + theme(axis.title.x = element_blank()),
plotlist[[7]] + annotation_custom(grid::textGrob("(g)", 0.08, 0.92)) + theme(axis.title.x = element_blank(), plot.margin = unit(c(10, 5.5, 5.5, 5.5), "points")),
plotlist[[8]] + annotation_custom(grid::textGrob("(h)", 0.08, 0.92)) + theme(axis.title.x = element_blank()),
plotlist[[9]] + annotation_custom(grid::textGrob("(i)", 0.15, 0.92)) + theme(axis.title.x = element_blank()),
plotlist[[10]] + annotation_custom(grid::textGrob("(j)", 0.08, 0.92)) + theme(axis.title.x = element_blank()),
plotlist[[11]] + annotation_custom(grid::textGrob("(k)", 0.08, 0.92)) + theme(axis.title.x = element_blank()),
plotlist[[12]] + annotation_custom(grid::textGrob("(l)", 0.1, 0.92)) + theme(axis.title.x = element_blank()),
nrow = 3, ncol = 4, byrow = FALSE, bottom = "Year")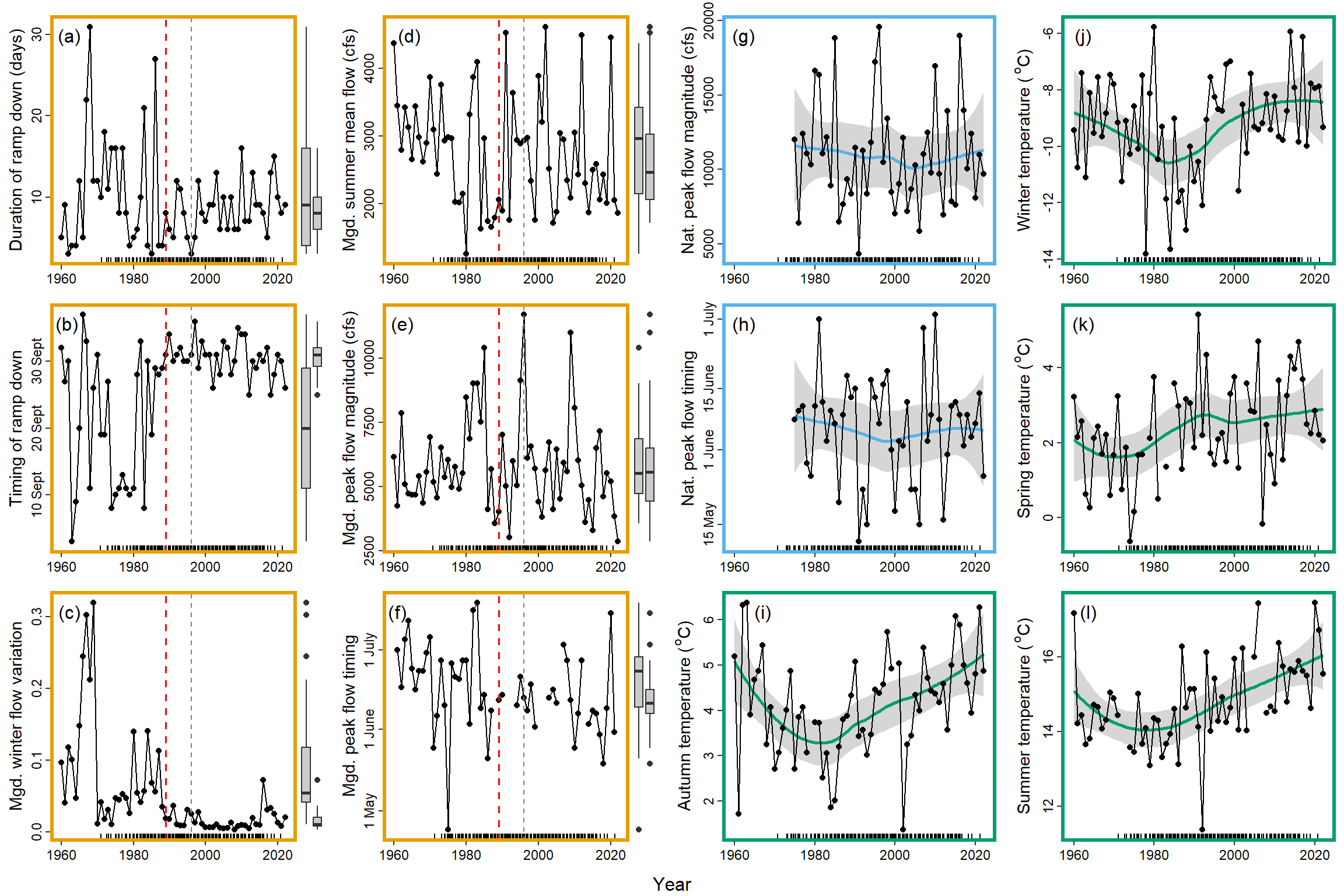
jpeg("Figures/Covariates/Covariates_TimeSeriesData_Combined.jpg", units = "in", width = 12, height = 8, res = 1500)
cp
dev.off()png
2