Code
siteinfo <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Data/EcoDrought_SiteInformation.csv")
siteinfo_sp <- st_as_sf(siteinfo, coords = c("long", "lat"), crs = 4326)Purpose: Use boxes to highlight additional details of the data, vignettes, and case studies that demonstrate spatiotemporal streamflow heterogeneity
Site information
siteinfo <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Data/EcoDrought_SiteInformation.csv")
siteinfo_sp <- st_as_sf(siteinfo, coords = c("long", "lat"), crs = 4326)Little g’s
dat_clean <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/LittleG_data_clean.csv")Big G’s
dat_clean_big <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/BigG_data_clean.csv")Climate
climdf <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/Daymet_climate.csv")
climdf_summ <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/Daymet_climate_summary.csv")Water availability
wateravail <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/BigG_wateravailability_annual.csv")Watersheds (filter to Snake River only)
sheds_list <- list()
myfiles <- list.files(path = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/", pattern = ".shp")
for (i in 1:length(myfiles)) {
sheds_list[[i]] <- st_read(paste("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/", myfiles[i], sep = ""))
}
sheds <- do.call(rbind, sheds_list) %>%
mutate(site_id = ifelse(site_id == "SP01", "SP07", ifelse(site_id == "SP07", "SP01", site_id))) %>%
left_join(siteinfo) %>%
filter(basin == "Snake River")
sheds <- st_transform(sheds, crs = crs(siteinfo_sp))
#mapview(sheds %>% arrange(desc(area_sqmi)), alpha.regions = 0.2)Spring prevalence
springprev <- rast("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Data/Snake Groundwater/SpringPrev_UpperSnake_BedSurf_nolakes_flowbuff100.tif")
plot(springprev)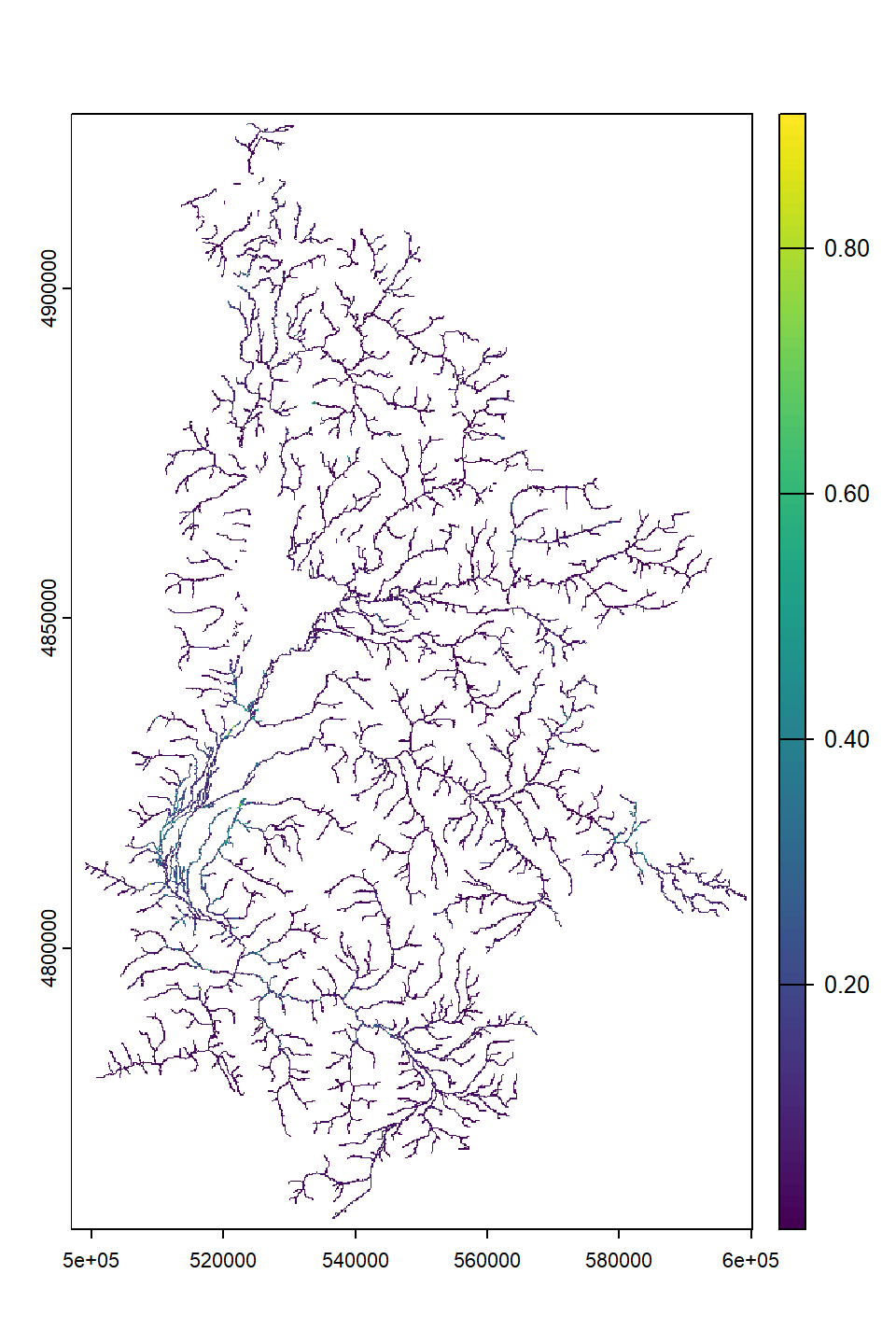
Purpose: Show what high-resolution temporal data (hourly) reveals about network diversity in streamflow response to individual storms (peak flow magnitude and timing and recession rates) and during low flow (diel fluctuations)
The Wedge Model for the West Brook
Purpose: At coarser time scales (summarized by event/baseflow periods), show how streamflow heterogeneity expands and contracts during wet to dry periods.
Purpose: Show what high-resolution spatial data reveals about network diversity in streamflow at a single point in time.
Purpose: Explore the effect of groundwater on relative summer (July-September) water availability.
Load PASTA daily derived parameters: summarize as July-September site-specific means (across all years)
pasta <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Covariates/pasta_derived_parameters_daily.csv") %>%
mutate(Month = month(date)) %>%
rename(CalendarYear = year) %>%
filter(Month %in% c(7:9)) %>%
group_by(site_name) %>%
summarize(meanRatio = mean(meanRatio, na.rm = TRUE),
phaseLag = mean(phaseLag, na.rm = TRUE),
amplitudeRatio = mean(amplitudeRatio, na.rm = TRUE))
pasta# A tibble: 73 × 4
site_name meanRatio phaseLag amplitudeRatio
<chr> <dbl> <dbl> <dbl>
1 Avery Brook 0.891 2.31 0.264
2 BigCreekLower 0.936 2.22 0.376
3 BigCreekMiddle 0.840 1.29 0.411
4 BigCreekUpper 0.664 2.31 0.179
5 Buck Creek 0.699 1.19 0.353
6 CoalCreekHeadwaters 0.670 2.11 0.209
7 CoalCreekLower 0.843 2.91 0.503
8 CoalCreekMiddle 0.692 1.27 0.223
9 CoalCreekNorth 0.733 2.49 0.246
10 Crandall Creek 0.758 1.76 0.612
# ℹ 63 more rowsCreate flow by groundwater plotting function.
mdaystib <- tibble(Month = c(1:12), mdays = c(31,28,31,30,31,30,31,31,30,31,30,31))
gwflowfun <- function (subbas, years, dropsites, months = c(7:9)) {
dat_clean %>%
filter(subbasin == subbas, CalendarYear %in% years, Month %in% months) %>%
group_by(site_name, subbasin, designation, CalendarYear) %>% #, Month, MonthName) %>%
summarise(ss = n(),
logYield = mean(logYield, na.rm = TRUE)) %>%
ungroup() %>%
#left_join(mdaystib) %>%
mutate(pdays = ss/92#,
#YearMonth = paste(CalendarYear, "_", Month, sep = "")
) %>%
filter(pdays > 0.9,
!site_name %in% dropsites) %>%
group_by(CalendarYear) %>%
mutate(z_logYield = scale(logYield, center = TRUE, scale = TRUE)[,1]) %>%
ungroup() %>%
left_join(pasta) %>%
ggplot(aes(x = amplitudeRatio, y = z_logYield)) +
geom_abline(intercept = 0, slope = 0, linetype = 2) +
geom_smooth(method = "lm", color = "black") +
geom_point(aes(color = site_name)) +
facet_wrap(~CalendarYear, nrow = 1) +
#facet_wrap2(~CalendarYear, nrow = 1, ncol = 5, trim_blank = FALSE) +
#facet_grid(cols = vars(Month), rows = vars(CalendarYear)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
}Plot the relationship between standardized annual summer mean discharge and amplitude ratio from PASTA, where lower amplitude ratio values are indicative of greater groundwater availability. Mean flow for each site is standardized by year to remove interannual variation in climate/regional water availability
gwflowfun(subbas = "West Brook", dropsites = c("West Brook Reservoir", "Mitchell Brook"), years = c(2020:2024))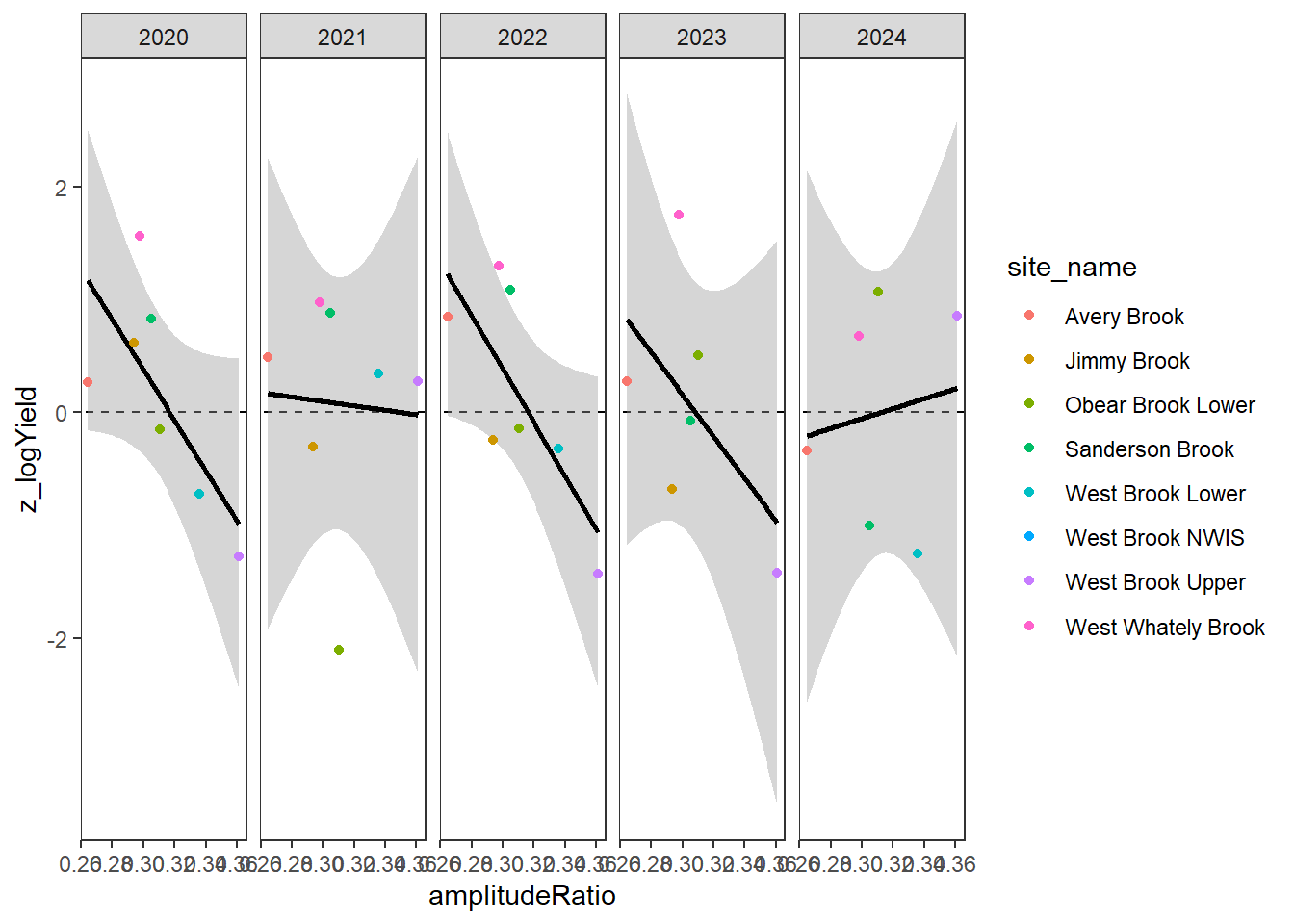
gwflowfun(subbas = "Staunton River", dropsites = NA, years = c(2019:2022))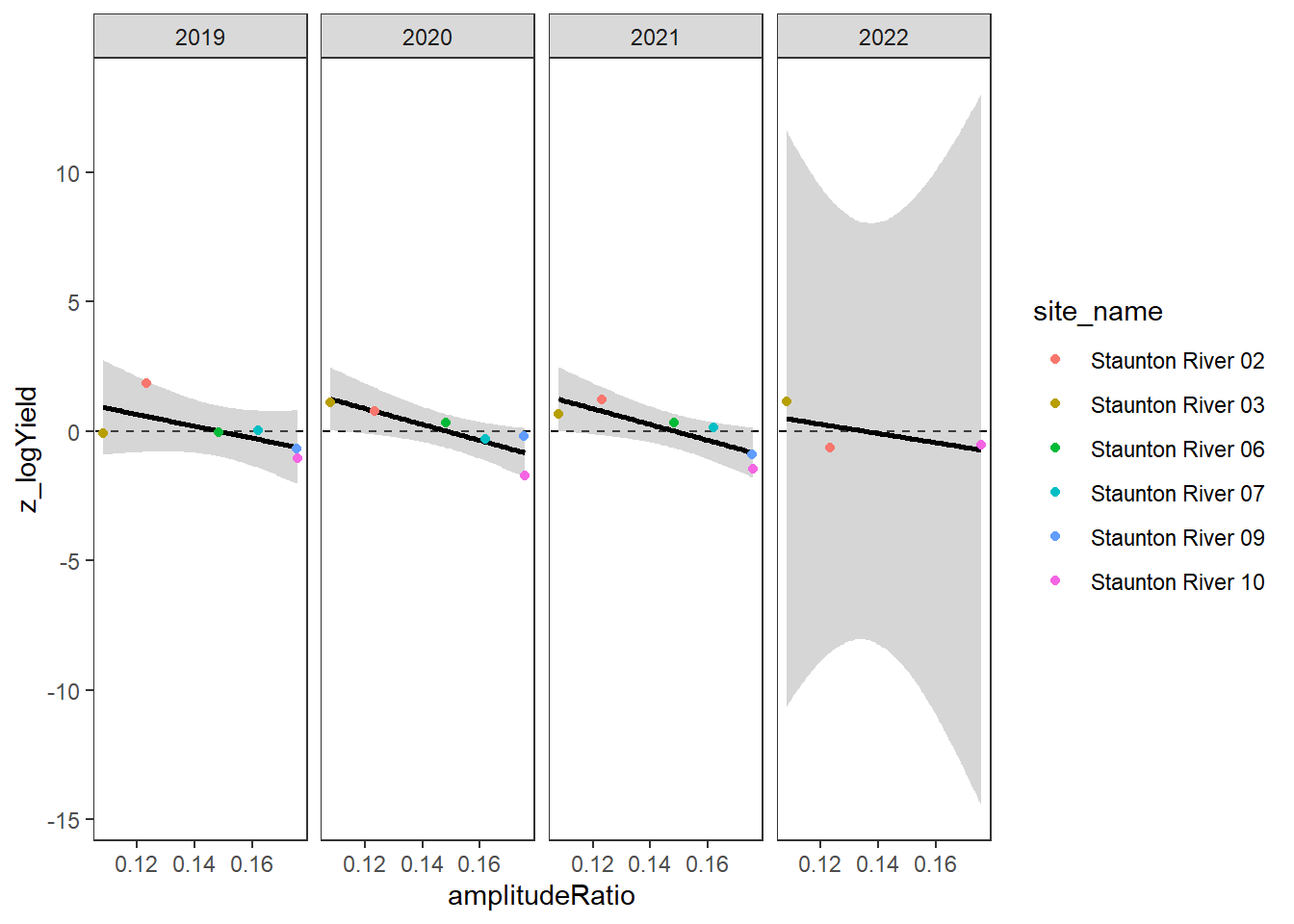
gwflowfun(subbas = "Snake River", dropsites = NA, years = c(2018, 2020:2022), months = c(7:9))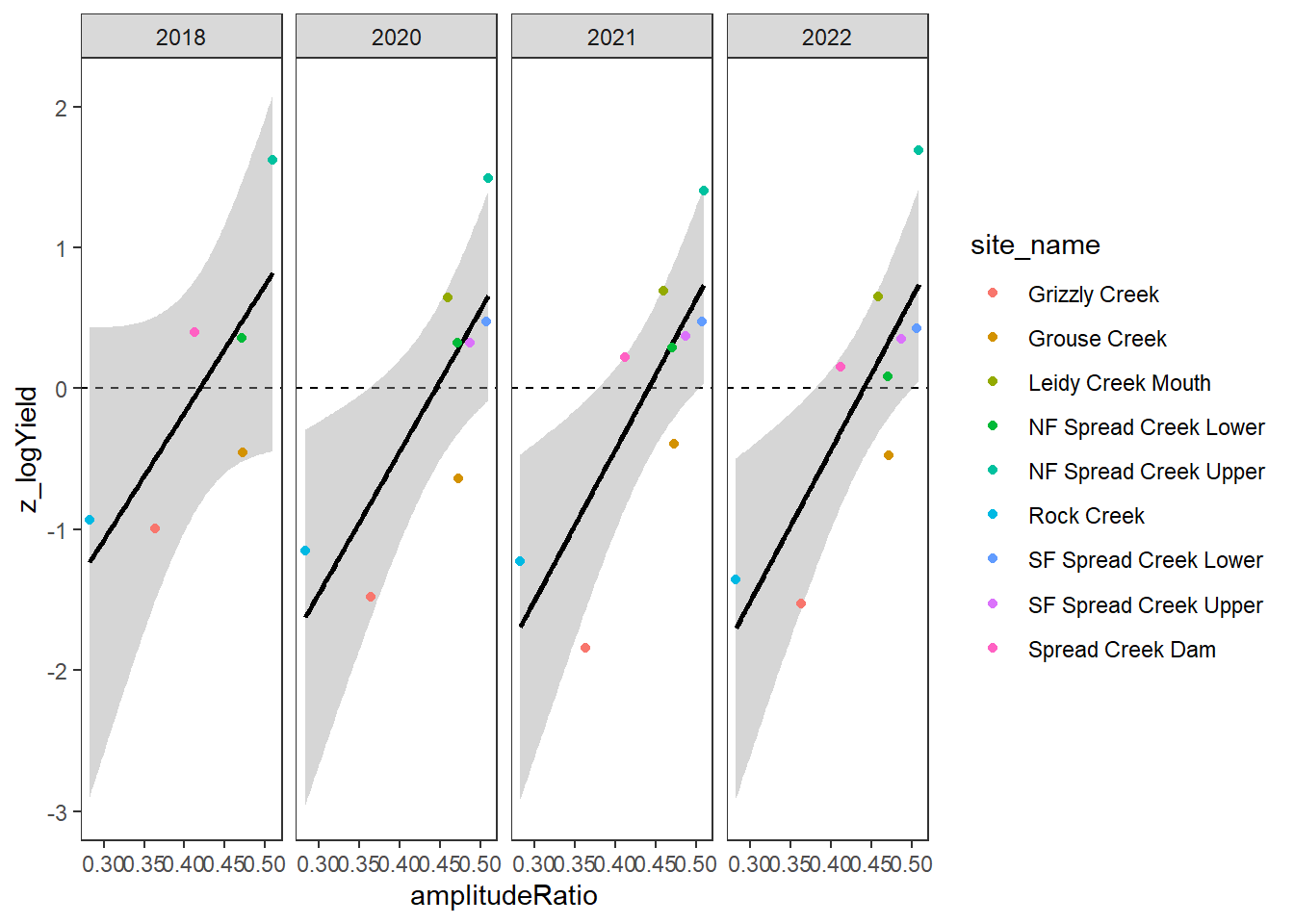
gwflowfun(subbas = "Shields River", dropsites = NA, years = c(2017,2019,2020,2022,2023), months = c(7:9))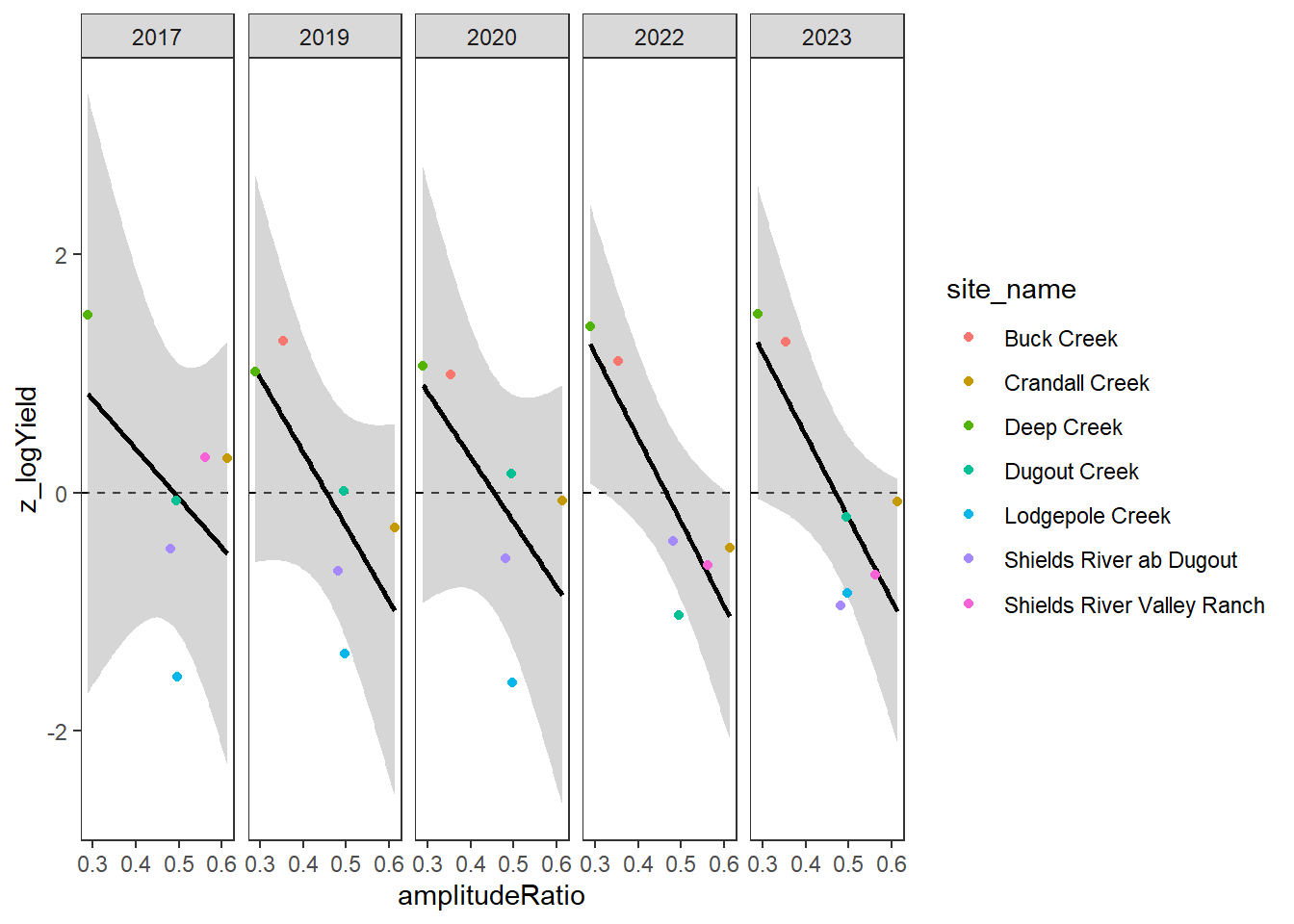
Extract groundwater metrics from MaxEnt spring prevalence model
# convert to SpatVectors
sites_flow <- vect(siteinfo_sp %>% filter(basin == "Snake River"))
sheds <- vect(sheds)
# transform
sites_flow <- terra::project(sites_flow, crs(springprev))
sheds <- terra::project(sheds, crs(springprev))
# extract average and weighted spring prevalence for each basin
sites <- sheds$site_name
gwlist <- list()
st <- Sys.time()
for (i in 1:length(sites)) {
spring_mask <- mask(crop(springprev, sheds[sheds$site_name == sites[i],]), sheds[sheds$site_name == sites[i],]) # crop and mask by basin
dist_rast <- distance(spring_mask, sites_flow[sites_flow$site_name == sites[i],]) %>% mask(spring_mask) # calculate distance between each raster cell and site location
gwlist[[i]] <- tibble(site_name = sites[i],
area_sqmi = sites_flow$area_sqmi[i],
springprev_point = terra::extract(springprev, sites_flow[sites_flow$site_name == sites[i],], na.rm = TRUE)[,2],
springprev_basinmean = as.numeric(global(spring_mask, "mean", na.rm = T)), # extract(spring_buff, sheds_yoy[sheds_yoy$site == sites[i],], fun = mean, na.rm = TRUE)[,2],
springprev_iew01km = as.numeric(global(spring_mask * (1 / exp(dist_rast/1000)), "sum", na.rm = T) / global(1 / exp(dist_rast/1000), "sum", na.rm = T)),
springprev_iew05km = as.numeric(global(spring_mask * (1 / exp(dist_rast/5000)), "sum", na.rm = T) / global(1 / exp(dist_rast/5000), "sum", na.rm = T))
)
print(i)
}
|---------|---------|---------|---------|
=========================================
[1] 1
|---------|---------|---------|---------|
=========================================
[1] 2
|---------|---------|---------|---------|
=========================================
[1] 3
|---------|---------|---------|---------|
=========================================
[1] 4
|---------|---------|---------|---------|
=========================================
[1] 5
|---------|---------|---------|---------|
=========================================
[1] 6
|---------|---------|---------|---------|
=========================================
[1] 7
|---------|---------|---------|---------|
=========================================
[1] 8
|---------|---------|---------|---------|
=========================================
[1] 9
|---------|---------|---------|---------|
=========================================
[1] 10
|---------|---------|---------|---------|
=========================================
[1] 11
|---------|---------|---------|---------|
=========================================
[1] 12
|---------|---------|---------|---------|
=========================================
[1] 13
|---------|---------|---------|---------|
=========================================
[1] 14Sys.time() - stTime difference of 7.001879 secsgwmetrics_snake <- do.call(rbind, gwlist) # bind as tibble
gwmetrics_snake# A tibble: 14 × 6
site_name area_sqmi springprev_point springprev_basinmean springprev_iew01km
<chr> <dbl> <dbl> <dbl> <dbl>
1 NF Spread… 12.7 0.0441 0.0502 0.0575
2 Grouse Cr… 5.15 0.00917 0.0140 0.0104
3 Leidy Cre… 5.17 0.00892 0.0320 0.0336
4 Leidy Cre… 2.18 0.0305 0.0503 0.0476
5 Leidy Cre… 5.16 0.0179 0.0324 0.0369
6 NF Spread… 27.9 0.0362 0.0402 0.0223
7 Grizzly C… 3.68 0.0379 0.0376 0.0333
8 Rock Creek 4.74 0.0175 0.00675 0.00830
9 SF Spread… 44.3 0.0277 0.0220 0.0119
10 SF Spread… 35.1 0.00378 0.0245 0.0253
11 Spread Cr… 97.5 0.0387 0.0286 0.0122
12 Pacific C… 166. 0.0771 0.0267 0.0442
13 Leidy Cre… 5.17 0.00634 0.0321 0.0345
14 SF Spread… 44.2 0.0194 0.0221 0.0119
# ℹ 1 more variable: springprev_iew05km <dbl>Plot groundwater index across upper Snake River basin
springprev_cont <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Data/Snake Groundwater/GroundwaterMetrics_Normalized_PredPoints.shp")
mapview(st_as_sf(springprev_cont), zcol = "springpre3", col.regions = viridis(n = 100, direction = -1))