Code
siteinfo <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Data/EcoDrought_SiteInformation.csv")
siteinfo_sp <- st_as_sf(siteinfo, coords = c("long", "lat"), crs = 4326)Purpose: Evaluate the extent and magnitude of spatial and temporal variation in headwater streamflow regimes.
Approach:
Notes:
N/A
Site information
siteinfo <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Data/EcoDrought_SiteInformation.csv")
siteinfo_sp <- st_as_sf(siteinfo, coords = c("long", "lat"), crs = 4326)Little g’s
dat_clean <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/LittleG_data_clean.csv")Big G’s
dat_clean_big <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/BigG_data_clean.csv")Climate
climdf <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/Daymet_climate.csv")
climdf_summ <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/Daymet_climate_summary.csv")Water availability. In western, snowmelt-dominated basins, annual (water year) total yield is strongly related to summer total yield. But in eastern, rain-dominated basins, the relationship is much weaker, suggesting “faster” response to climate forcing in rain-dominated basins.
wateravail <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/BigG_wateravailability_annual.csv") %>%
filter(!is.na(totalyield), !is.na(totalyield_sum)) %>%
group_by(site_name) %>%
mutate(tyz_perc = percentile(totalyield_z),
tyz_sum_perc = percentile(totalyield_sum_z)) %>%
mutate(tyz_perc = ifelse(is.na(tyz_perc), 0, tyz_perc),
tyz_sum_perc = ifelse(is.na(tyz_sum_perc), 0, tyz_sum_perc))
wateravail %>%
filter(basin != "Piney River") %>%
mutate(basin = ifelse(basin == "Shields River", "Yellowstone River", basin)) %>%
mutate(basin = factor(basin, levels = c("West Brook", "Staunton River", "Paine Run", "Flathead", "Yellowstone River", "Snake River", "Donner Blitzen"))) %>%
ggplot(aes(x = tyz_perc, y = tyz_sum_perc)) +
geom_point() +
geom_smooth(method = "lm") +
xlab("Total annual yield (percentile)") + ylab("Total summer yield (percentile)") +
facet_wrap(~basin) + theme_bw()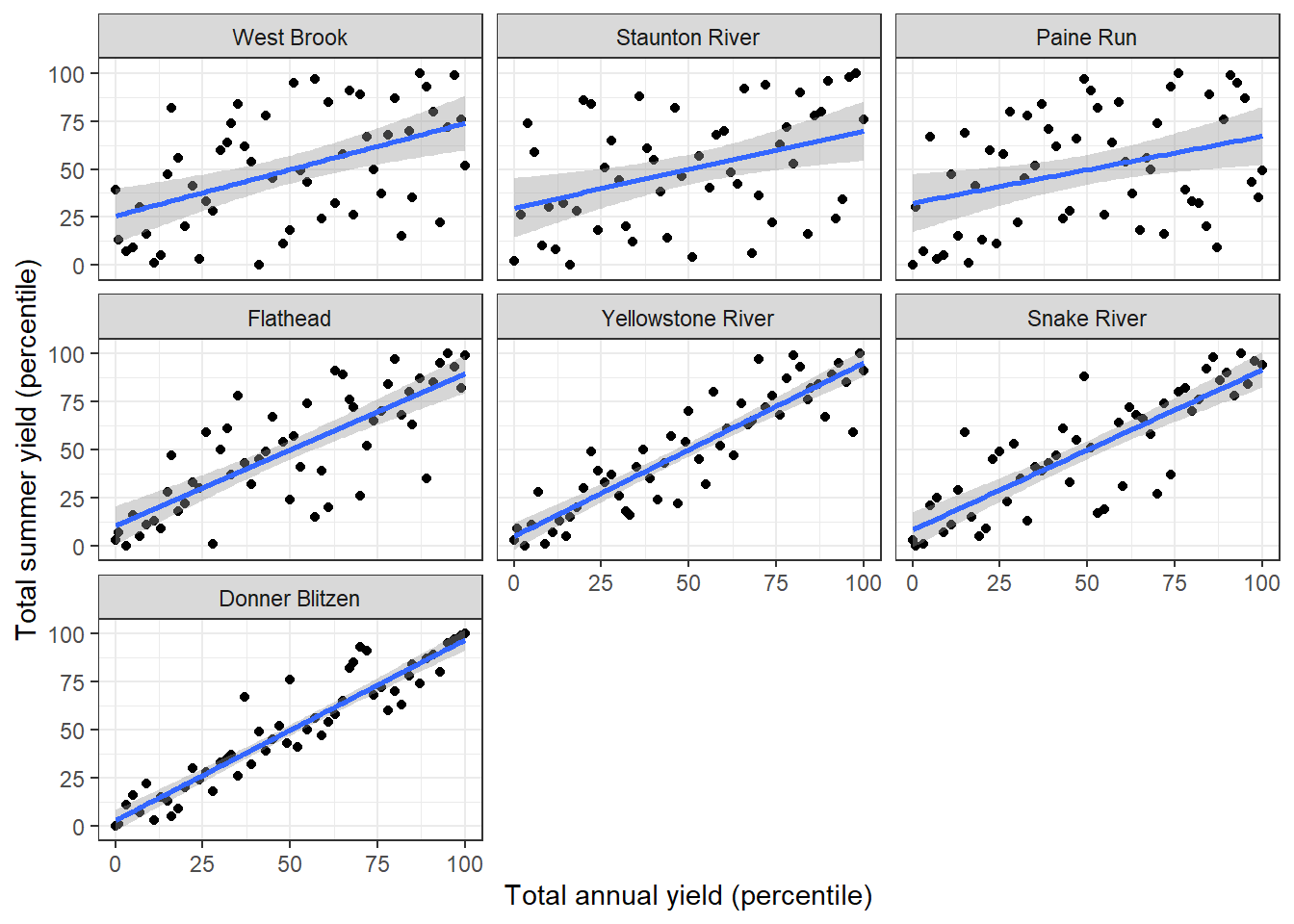
Watersheds
sheds_list <- list()
myfiles <- list.files(path = "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/", pattern = ".shp")
for (i in 1:length(myfiles)) {
sheds_list[[i]] <- st_read(paste("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/", myfiles[i], sep = ""))
}
sheds <- do.call(rbind, sheds_list) %>%
mutate(site_id = ifelse(site_id == "SP01", "SP07", ifelse(site_id == "SP07", "SP01", site_id))) %>%
left_join(siteinfo)
#mapview(sheds %>% arrange(desc(area_sqmi)), alpha.regions = 0.2)For colors, order sites from downstream to upstream (roughly) and by subbasin (if appropriate)
wborder <- c("West Brook NWIS", "West Brook Lower", "Mitchell Brook", "Jimmy Brook", "Obear Brook Lower", "West Brook Upper", "West Brook Reservoir", "Sanderson Brook", "Avery Brook", "West Whately Brook")
paineorder <- c("Paine Run 10", "Paine Run 08", "Paine Run 07", "Paine Run 06", "Paine Run 02", "Paine Run 01")
stauntorder <- c("Staunton River 10", "Staunton River 09", "Staunton River 07", "Staunton River 06", "Staunton River 03", "Staunton River 02")
flatorder <- c("BigCreekLower", "LangfordCreekLower", "LangfordCreekUpper", "Big Creek NWIS", "BigCreekUpper", "HallowattCreekLower", "NicolaCreek", "WernerCreek", "Hallowat Creek NWIS", "CoalCreekLower", "CycloneCreekLower", "CycloneCreekMiddle", "CycloneCreekUpper", "CoalCreekMiddle", "CoalCreekNorth", "CoalCreekHeadwaters", "McGeeCreekLower", "McGeeCreekTrib", "McGeeCreekUpper")
yellorder <- c("Shields River Valley Ranch", "Deep Creek", "Crandall Creek", "Buck Creek", "Dugout Creek", "Shields River ab Dugout", "Lodgepole Creek", "EF Duck Creek be HF", "EF Duck Creek ab HF", "Henrys Fork")
snakeorder <- c("Spread Creek Dam", "Rock Creek", "NF Spread Creek Lower", "NF Spread Creek Upper", "Grizzly Creek", "SF Spread Creek Lower", "Grouse Creek", "SF Spread Creek Upper", "Leidy Creek Mouth")
donnerorder <- c("Fish Creek NWIS", "Donner Blitzen ab Fish NWIS", "Donner Blitzen nr Burnt Car NWIS", "Donner Blitzen ab Indian NWIS")Currently not being used…
View daily time series data by sub-basin. Note that we are using the “Super G” NWIS data for the reference gage (black line). Per Robert comment, entirely nested design is cute, but doesn’t reflect how the data is actually used.
Big G NWIS sites/reference gages for each basin/subbasin:
dat_clean_big %>% group_by(region, basin, subbasin) %>% summarize(site_name = unique(site_name)) %>% ungroup() %>% filter(!is.na(region)) %>% kable()| region | basin | subbasin | site_name |
|---|---|---|---|
| Flat | Flathead | Flathead | North Fork Flathead River NWIS |
| Mass | West Brook | West Brook | South River Conway NWIS |
| Oreg | Donner Blitzen | Donner Blitzen | Donner Blitzen River nr Frenchglen NWIS |
| Shen | Paine Run | Paine Run | South River Harriston NWIS |
| Shen | Piney River | Piney River | Battle Run NWIS |
| Shen | Staunton River | Staunton River | Rapidan River NWIS |
| Shields | Shields River | Shields River | Yellowstone River Livingston NWIS |
| Snake | Snake River | Snake River | Pacific Creek at Moran NWIS |
dat_clean %>% group_by(region, basin) %>% summarize(subbasin = unique(subbasin)) %>% ungroup() #%>% filter(!is.na(region)) %>% kable()# A tibble: 10 × 3
region basin subbasin
<chr> <chr> <chr>
1 Flat Flathead Big Creek
2 Flat Flathead Coal Creek
3 Flat Flathead McGee Creek
4 Mass West Brook West Brook
5 Oreg Donner Blitzen Donner Blitzen
6 Shen Paine Run Paine Run
7 Shen Staunton River Staunton River
8 Shields Shields River Shields River
9 Shields Shields River Duck Creek
10 Snake Snake River Snake River dat_clean %>%
filter(basin == "Flathead") %>%
bind_rows(dat_clean_big %>% filter(basin == "Flathead", date >= date("2017-07-29"), date <= date("2023-12-11"))) %>%
select(date, site_name, logYield) %>%
spread(key = site_name, value = logYield) %>%
relocate("North Fork Flathead River NWIS", .after = last_col()) %>%
dygraph() %>% dyRangeSelector() %>% dyAxis("y", label = "ln(Yield, mm)") %>%
#dyOptions(colors = c(brewer.pal(7, "Dark2"), "black")) %>%
dySeries("North Fork Flathead River NWIS", strokeBorderWidth = 1, strokeWidth = 1.5, color = "black")dat_clean %>%
filter(basin == "Shields River") %>%
bind_rows(dat_clean_big %>% filter(basin == "Shields River", date >= date("2015-04-01"), date <= date("2023-12-31"))) %>%
select(date, site_name, logYield) %>%
spread(key = site_name, value = logYield) %>%
relocate("Yellowstone River Livingston NWIS", .after = last_col()) %>%
dygraph() %>% dyRangeSelector() %>% dyAxis("y", label = "ln(Yield, mm)") %>%
#dyOptions(colors = c(brewer.pal(7, "Dark2"), "black")) %>%
dySeries("Yellowstone River Livingston NWIS", strokeBorderWidth = 1, strokeWidth = 1.5, color = "black")Create time series data plots objects
polycol <- "grey90"
mycols <- brewer.pal(7, "Set2")
### WEST BROOK
tempdat <- dat_clean %>%
filter(basin == "West Brook") %>%
mutate(site_name = factor(site_name, levels = wborder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "West Brook", date >= date("2020-01-01"), date <= date("2025-01-03"))
# color
pwb1 <- ggplot() +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2023-07-01"), xmax = ymd("2023-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2024-07-01"), xmax = ymd("2024-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = wborder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
ylim(-3,2) +
with_outer_glow(annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "West Brook, MA", hjust = 0, vjust = 1), colour = "white", expand = 10) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[1], fill = NA, linewidth = 1.5)
)
# greyscale
pwb2 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, group = site_name), color = "grey") +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "black", size = 1) +
ylim(-3,2) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "West Brook, MA", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### STAUNTON RIVER
tempdat <- dat_clean %>%
filter(basin == "Staunton River") %>%
mutate(site_name = factor(site_name, levels = stauntorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Staunton River", date >= as_date("2018-11-07"), date <= as_date("2022-10-19"))
# color
pst1 <- ggplot() +
geom_rect(aes(xmin = ymd("2019-07-01"), xmax = ymd("2019-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = stauntorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
with_outer_glow(annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Staunton River, VA", hjust = 0, vjust = 1), colour = "white", expand = 10) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[2], fill = NA, linewidth = 1.5)
)
# greyscale
pst2 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, group = site_name), color = "grey") +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "black", size = 1) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Staunton River", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black"), panel.border = element_rect(colour = mycols[2], fill = NA, linewidth = 2))
### PAINE RUN
tempdat <- dat_clean %>%
filter(basin == "Paine Run") %>%
mutate(site_name = factor(site_name, levels = paineorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Paine Run", date >= as_date("2018-11-07"), date <= as_date("2023-05-15"))
# color
ppa1 <- ggplot() +
geom_rect(aes(xmin = ymd("2019-07-01"), xmax = ymd("2019-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = paineorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
with_outer_glow(annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Paine Run, VA", hjust = 0, vjust = 1), colour = "white", expand = 10) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[3], fill = NA, linewidth = 1.5)
)
# greyscale
ppa2 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, group = site_name), color = "grey") +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "black", size = 1) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Paine Run", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### FLATHEAD
tempdat <- dat_clean %>%
filter(basin == "Flathead", date >= date("2018-07-29"), date <= date("2023-08-11")) %>%
mutate(site_name = factor(site_name, levels = flatorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Flathead", date >= date("2018-07-29"), date <= date("2023-08-11"))
# color
pfl1 <- ggplot() +
geom_rect(aes(xmin = ymd("2018-07-29"), xmax = ymd("2018-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2019-07-01"), xmax = ymd("2019-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2023-07-01"), xmax = ymd("2023-08-11"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = flatorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
with_outer_glow(annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Flathead River, MT", hjust = 0, vjust = 1), colour = "white", expand = 10) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[4], fill = NA, linewidth = 1.5)
)
# greyscale
pfl2 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, group = site_name), color = "grey") +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "black", size = 1) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Flathead River", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### YELLOWSTONE
tempdat <- dat_clean %>%
filter(basin == "Shields River", date >= date("2016-06-01"), date <= date("2023-11-01")) %>%
mutate(site_name = factor(site_name, levels = yellorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Shields River", date >= date("2016-06-01"), date <= date("2023-11-01"))
# color
pye1 <- ggplot() +
geom_rect(aes(xmin = ymd("2016-07-01"), xmax = ymd("2016-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2017-07-01"), xmax = ymd("2017-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2018-07-01"), xmax = ymd("2018-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2019-07-01"), xmax = ymd("2019-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2023-07-01"), xmax = ymd("2023-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = yellorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
with_outer_glow(annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Yellowstone River, MT", hjust = 0, vjust = 1), colour = "white", expand = 10) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[5], fill = NA, linewidth = 1.5)
)
# greyscale
pye2 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, group = site_name), color = "grey") +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "black", size = 1) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Yellowstone River, MT", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### SNAKE
tempdat <- dat_clean %>%
filter(basin == "Snake River", date >= date("2016-06-01"), date <= date("2023-10-03")) %>%
mutate(site_name = factor(site_name, levels = snakeorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Snake River", date >= date("2016-06-01"), date <= date("2023-10-03"))
# color
psn1 <- ggplot() +
geom_rect(aes(xmin = ymd("2016-07-01"), xmax = ymd("2016-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2017-07-01"), xmax = ymd("2017-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2018-07-01"), xmax = ymd("2018-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2019-07-01"), xmax = ymd("2019-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2023-07-01"), xmax = ymd("2023-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = snakeorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
with_outer_glow(annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Snake River, WY", hjust = 0, vjust = 1), colour = "white", expand = 10) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[6], fill = NA, linewidth = 1.5)
)
# greyscale
psn2 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, group = site_name), color = "grey") +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "black", size = 1) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Snake River, WY", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
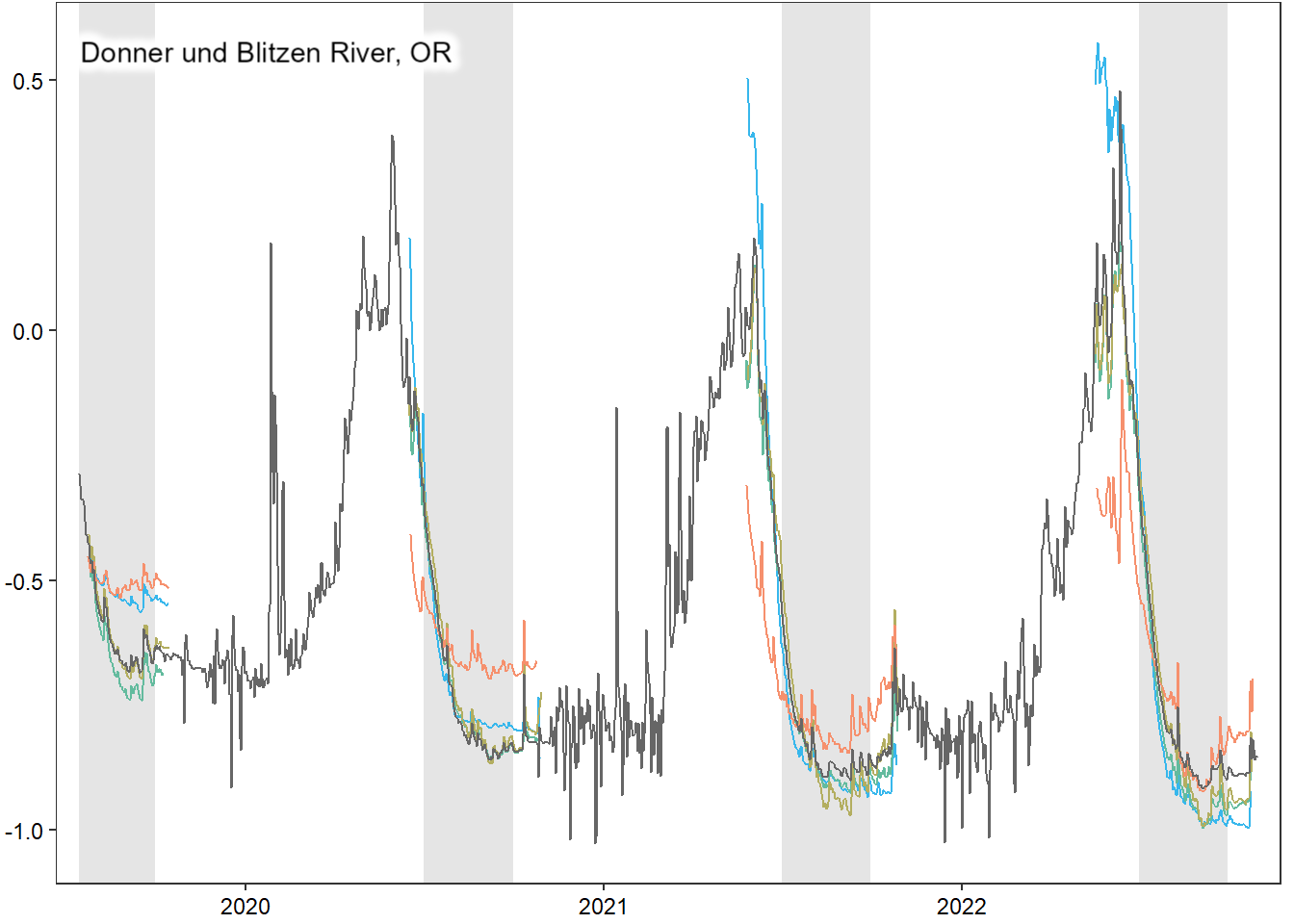
### DONNER BLITZEN
tempdat <- dat_clean %>%
filter(basin == "Donner Blitzen") %>%
mutate(site_name = factor(site_name, levels = donnerorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Donner Blitzen", date >= date("2019-07-15"), date <= as_date("2022-10-30"))
# color
pdb1 <- ggplot() +
geom_rect(aes(xmin = ymd("2019-07-15"), xmax = ymd("2019-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = donnerorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
with_outer_glow(annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Donner und Blitzen River, OR", hjust = 0, vjust = 1), colour = "white", expand = 10) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[7], fill = NA, linewidth = 1.5)
)
# greyscale
pdb2 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, group = site_name), color = "grey") +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "black", size = 1) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Donner und Blitzen River", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))ggplot() +
geom_rect(aes(xmin = ymd("2019-07-15"), xmax = ymd("2019-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2020-07-01"), xmax = ymd("2020-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2021-07-01"), xmax = ymd("2021-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_rect(aes(xmin = ymd("2022-07-01"), xmax = ymd("2022-09-30"), ymin = -Inf, ymax = Inf), fill = polycol, color = NA) +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = donnerorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
with_outer_glow(annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Donner und Blitzen River, OR", hjust = 0, vjust = 1), colour = "white", expand = 10) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[7], fill = NA, linewidth = 1.5)
)
Generate plot
annotate_figure(egg::ggarrange(pwb1, pst1, ppa1, pfl1, pye1, psn1, pdb1, ncol = 1), left = "log(Yield, mm/day)")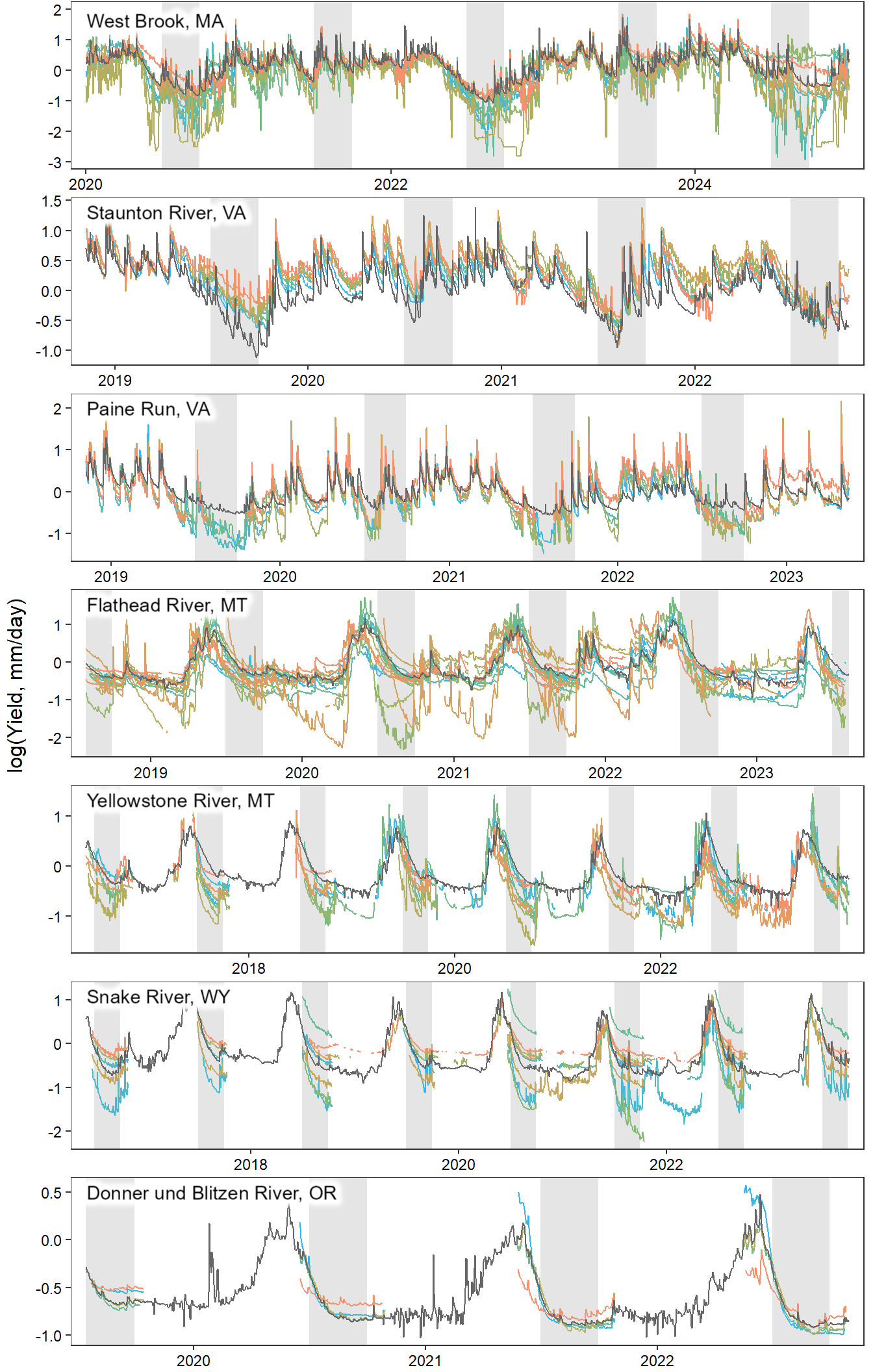
Write to file
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_timeseries_new_noborder.jpg", width = 5, height = 11, units = "in", res = 1000)
annotate_figure(egg::ggarrange(pwb1, pst1, ppa1, pfl1, pye1, psn1, pdb1, ncol = 1), left = "log(Yield, mm/day)")
dev.off()png
2 Plot time series just for summer 2020, for more detailed view
polycol <- "grey90"
mycols <- brewer.pal(7, "Set2")
### WEST BROOK
tempdat <- dat_clean %>%
filter(basin == "West Brook", date >= date("2020-07-01"), date <= date("2020-09-30")) %>%
mutate(site_name = factor(site_name, levels = wborder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "West Brook", date >= date("2020-07-01"), date <= date("2020-09-30"))
# same yaxis scale as main time series
pwb1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = wborder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 7, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[1], fill = NA, linewidth = 1.5)
)
### STAUNTON RIVER
tempdat <- dat_clean %>%
filter(basin == "Staunton River", date >= date("2020-07-01"), date <= date("2020-09-30")) %>%
mutate(site_name = factor(site_name, levels = stauntorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Staunton River", , date >= date("2020-07-01"), date <= date("2020-09-30"))
# color
pst1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = stauntorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 7, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[2], fill = NA, linewidth = 1.5)
)
### PAINE RUN
tempdat <- dat_clean %>%
filter(basin == "Paine Run", date >= date("2020-07-01"), date <= date("2020-09-30")) %>%
mutate(site_name = factor(site_name, levels = paineorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Paine Run", date >= date("2020-07-01"), date <= date("2020-09-30"))
# color
ppa1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = paineorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 7, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[3], fill = NA, linewidth = 1.5)
)
### FLATHEAD
tempdat <- dat_clean %>%
filter(basin == "Flathead", date >= date("2020-07-01"), date <= date("2020-09-30")) %>%
mutate(site_name = factor(site_name, levels = flatorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Flathead", date >= date("2020-07-01"), date <= date("2020-09-30"))
# color
pfl1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = flatorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 7, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[4], fill = NA, linewidth = 1.5)
)
### YELLOWSTONE
tempdat <- dat_clean %>%
filter(basin == "Shields River", date >= date("2020-07-01"), date <= date("2020-09-30")) %>%
mutate(site_name = factor(site_name, levels = yellorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Shields River", date >= date("2020-07-01"), date <= date("2020-09-30"))
# color
pye1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = yellorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 7, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[5], fill = NA, linewidth = 1.5)
)
### SNAKE
tempdat <- dat_clean %>%
filter(basin == "Snake River", date >= date("2020-07-01"), date <= date("2020-09-30")) %>%
mutate(site_name = factor(site_name, levels = snakeorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Snake River", date >= date("2020-07-01"), date <= date("2020-09-30"))
# color
psn1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = snakeorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 7, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[6], fill = NA, linewidth = 1.5)
)
### DONNER BLITZEN
tempdat <- dat_clean %>%
filter(basin == "Donner Blitzen", date >= date("2020-07-01"), date <= date("2020-09-30")) %>%
mutate(site_name = factor(site_name, levels = donnerorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Donner Blitzen", date >= date("2020-07-01"), date <= date("2020-09-30"))
# color
pdb1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = donnerorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 7, b = 2, l = 2),
axis.text = element_text(color = "black")#, panel.border = element_rect(colour = mycols[7], fill = NA, linewidth = 1.5)
)Write to file
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_timeseries_2020only.jpg", width = 2, height = 11, units = "in", res = 1000)
annotate_figure(egg::ggarrange(pwb1, pst1, ppa1, pfl1, pye1, psn1, pdb1, ncol = 1))
dev.off()png
2 How does the range of (summer) flow change with water availability?
First, on daily values: (NOT EVALUATED)
tempdat <- dat_clean %>%
filter(basin == "West Brook", !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = wborder))
tempdat <- tempdat %>% left_join(dat_clean_big %>% filter(basin == "West Brook") %>% select(basin, date, logYield) %>% rename(logYield_big = logYield))
tempdat <- tempdat %>% group_by(date) %>% summarize(nsites = n(), lgrange = range(logYield)[2] - range(logYield)[1], logYield_big = unique(logYield_big))
p1 <- tempdat %>% filter(nsites == 10) %>%
ggplot(aes(x = logYield_big, y = log(lgrange), group = year(date))) +
geom_point() +
geom_smooth(method = "lm") +
ggtitle("West Brook") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.text = element_text(color = "black"), legend.position = "none")
tempdat <- dat_clean %>%
filter(basin == "Paine Run", !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = wborder))
tempdat <- tempdat %>% left_join(dat_clean_big %>% filter(basin == "Paine Run") %>% select(basin, date, logYield) %>% rename(logYield_big = logYield))
tempdat <- tempdat %>% group_by(date) %>% summarize(nsites = n(), lgrange = range(logYield)[2] - range(logYield)[1], logYield_big = unique(logYield_big))
p2 <- tempdat %>% filter(nsites == 5) %>%
ggplot(aes(x = logYield_big, y = log(lgrange), group = year(date))) +
geom_point() +
geom_smooth(method = "lm") +
ggtitle("Paine Run") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.text = element_text(color = "black"), legend.position = "none")
tempdat <- dat_clean %>%
filter(basin == "Staunton River", !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = wborder))
tempdat <- tempdat %>% left_join(dat_clean_big %>% filter(basin == "Staunton River") %>% select(basin, date, logYield) %>% rename(logYield_big = logYield))
tempdat <- tempdat %>% group_by(date) %>% summarize(nsites = n(), lgrange = range(logYield)[2] - range(logYield)[1], logYield_big = unique(logYield_big))
p3 <- tempdat %>% filter(nsites == 6) %>%
ggplot(aes(x = logYield_big, y = log(lgrange), group = year(date))) +
geom_point() +
geom_smooth(method = "lm") +
ggtitle("Staunton River") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.text = element_text(color = "black"), legend.position = "none")
tempdat <- dat_clean %>%
filter(basin == "Flathead", !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = wborder))
tempdat <- tempdat %>% left_join(dat_clean_big %>% filter(basin == "Flathead") %>% select(basin, date, logYield) %>% rename(logYield_big = logYield))
tempdat <- tempdat %>% group_by(date) %>% summarize(nsites = n(), lgrange = range(logYield)[2] - range(logYield)[1], logYield_big = unique(logYield_big))
p4 <- tempdat %>% filter(nsites >= 10) %>%
ggplot(aes(x = logYield_big, y = log(lgrange), group = year(date))) +
geom_point() +
geom_smooth(method = "lm") +
ggtitle("Flathead River") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.text = element_text(color = "black"), legend.position = "none")
tempdat <- dat_clean %>%
filter(basin == "Shields River", !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = wborder))
tempdat <- tempdat %>% left_join(dat_clean_big %>% filter(basin == "Shields River") %>% select(basin, date, logYield) %>% rename(logYield_big = logYield))
tempdat <- tempdat %>% group_by(date) %>% summarize(nsites = n(), lgrange = range(logYield)[2] - range(logYield)[1], logYield_big = unique(logYield_big))
p5 <- tempdat %>% filter(nsites == 9) %>%
ggplot(aes(x = logYield_big, y = log(lgrange), group = year(date))) +
geom_point() +
geom_smooth() +
ggtitle("Yellowstone River") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.text = element_text(color = "black"), legend.position = "none")
tempdat <- dat_clean %>%
filter(basin == "Snake River", !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = wborder))
tempdat <- tempdat %>% left_join(dat_clean_big %>% filter(basin == "Snake River") %>% select(basin, date, logYield) %>% rename(logYield_big = logYield))
tempdat <- tempdat %>% group_by(date) %>% summarize(nsites = n(), lgrange = range(logYield)[2] - range(logYield)[1], logYield_big = unique(logYield_big))
p6 <- tempdat %>% filter(nsites == 9) %>%
ggplot(aes(x = logYield_big, y = log(lgrange), group = year(date))) +
geom_point() +
geom_smooth(method = "lm") +
ggtitle("Snake River") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.text = element_text(color = "black"), legend.position = "none")
tempdat <- dat_clean %>%
filter(basin == "Donner Blitzen", !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = wborder))
tempdat <- tempdat %>% left_join(dat_clean_big %>% filter(basin == "Donner Blitzen") %>% select(basin, date, logYield) %>% rename(logYield_big = logYield))
tempdat <- tempdat %>% group_by(date) %>% summarize(nsites = n(), lgrange = range(logYield)[2] - range(logYield)[1], logYield_big = unique(logYield_big))
p7 <- tempdat %>% filter(nsites == 4) %>%
ggplot(aes(x = logYield_big, y = log(lgrange), group = year(date))) +
geom_point() +
geom_smooth(method = "lm") +
ggtitle("Donner Blitzen") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.text = element_text(color = "black"), legend.position = "none")
annotate_figure(egg::ggarrange(p1 + theme(axis.title.x = element_blank(), axis.title.y = element_blank()),
p2 + theme(axis.title.x = element_blank(), axis.title.y = element_blank()),
p3 + theme(axis.title.x = element_blank(), axis.title.y = element_blank()),
p4 + theme(axis.title.x = element_blank(), axis.title.y = element_blank()),
p5 + theme(axis.title.x = element_blank(), axis.title.y = element_blank()),
p6 + theme(axis.title.x = element_blank(), axis.title.y = element_blank()),
p7 + theme(axis.title.x = element_blank(), axis.title.y = element_blank())),
bottom = "log(Yield at big G)", left = "Range of little g log(Yield) (logged)")Create function to plot interannual distribution of streamflow values, annual distributions of streamflow values, and scatterplot plot of
portfolioplot <- function(bas, orderr, type = c("interann", "annual", "scatter"), wtryrs) {
# filter data
tempdat <- dat_clean %>%
filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% wtryrs) %>%
mutate(site_name = factor(site_name, levels = orderr))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% unique(tempdat$WaterYear))
# calculate variability for reference gage
varbig <- tempdat_big %>%
group_by(WaterYear) %>%
summarize(bigrange = range(logYield)[2]-range(logYield)[1],
bigsd = sd(logYield),
bigvar = var(logYield)) %>%
ungroup()
# calculate total summer water availability from reference gage
#summerflow <- tempdat_big %>% group_by(WaterYear) %>% summarize(summeryield = sum(Yield_mm), summeryield_log = log(sum(Yield_mm))) %>% ungroup()
#wateravail2 <- wateravail %>% select(basin, WaterYear, totalyield, totalyield_z)
# calculate relative variation
pedat <- tempdat %>%
group_by(basin, WaterYear) %>%
summarize(nsites = length(unique(site_name)),
littlerange = range(logYield)[2]-range(logYield)[1],
littlesd = sd(logYield),
littlevar = var(logYield)) %>%
ungroup() %>%
left_join(varbig) %>%
mutate(pe_range = ((littlerange-bigrange)/bigrange),
pe_sd = ((littlesd-bigsd)/bigsd),
pe_var = ((littlevar-bigvar)/bigvar)) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield, totalyield_z, totalyield_sum, totalyield_sum_z, tyz_sum_perc)) %>%
filter(WaterYear %in% wtryrs) %>%
mutate(wylab = substr(WaterYear, 3, 4))
# interannual
pint <- ggplot() +
geom_density(data = tempdat, aes(x = logYield, y = ..scaled.., color = site_name, fill = site_name), size = 0.8) +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_fill_manual(values = alpha(cet_pal(nsites, name = "i1"), 0.1)) +
geom_density(data = tempdat_big, aes(x = logYield, y = ..scaled..), color = "grey40", fill = alpha("grey40", 0.2), size = 0.8) +
xlab("Summer log(Yield, mm/day)") + ylab("Scaled density") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.text = element_text(color = "black"), legend.position = "none")
# annual
pann <- ggplot() +
geom_density(data = tempdat, aes(x = logYield, y = ..scaled.., color = site_name, fill = site_name), size = 0.8) +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_fill_manual(values = alpha(cet_pal(nsites, name = "i1"), 0.1)) +
geom_density(data = tempdat_big, aes(x = logYield, y = ..scaled..), color = "grey40", fill = alpha("grey40", 0.2), size = 0.8) +
xlab("Summer log(Yield, mm/day)") + ylab("Scaled density") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.text = element_text(color = "black"), legend.position = "none") +
facet_wrap(~factor(WaterYear, levels = as.numeric(unlist(pedat %>% arrange(totalyield_sum_z) %>% select(WaterYear)))))
# portfolio effect by water availability
ppew <- pedat %>%
ggplot(aes(x = tyz_sum_perc, y = pe_range, label = wylab)) +
geom_abline(slope = 0, intercept = 0, linetype = 2) +
geom_smooth(method = "lm", color = "black") +
geom_point(shape = 21, fill = "skyblue1", size = 3) +
geom_text(vjust = -0.7, color = "grey40") +
xlab("Water availability") + ylab("Relative difference in range") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), axis.text = element_text(color = "black")) +
guides(color = guide_legend(title="Water year")) + xlim(0,100) + ylim(-1,4.5)
if(type == "interannual") { return(pint) }
if(type == "annual") { return(pann) }
if(type == "scatter") { return(ppew) }
if(type == "table") { return(pedat) }
}Generate plots
denwb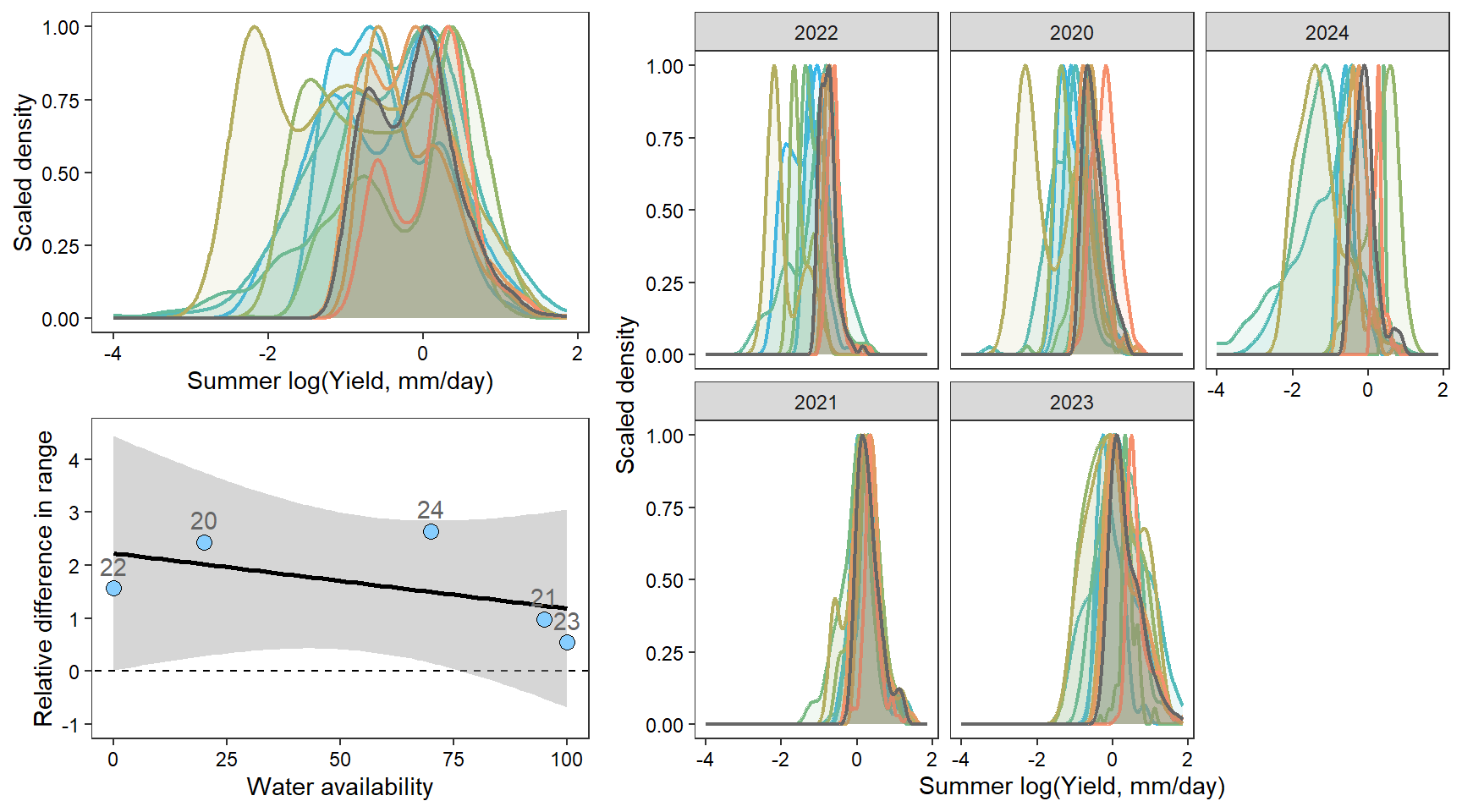
denpa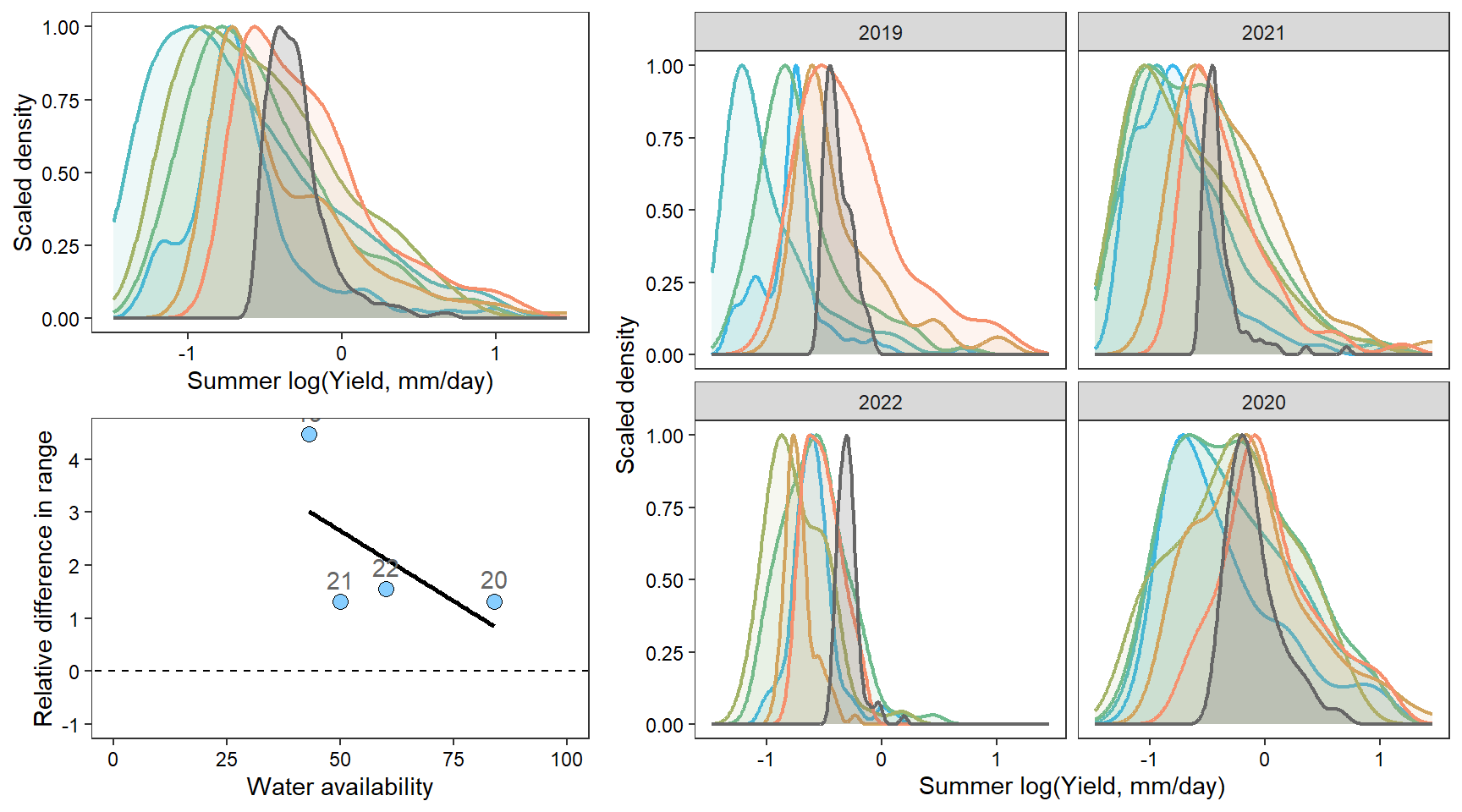
denst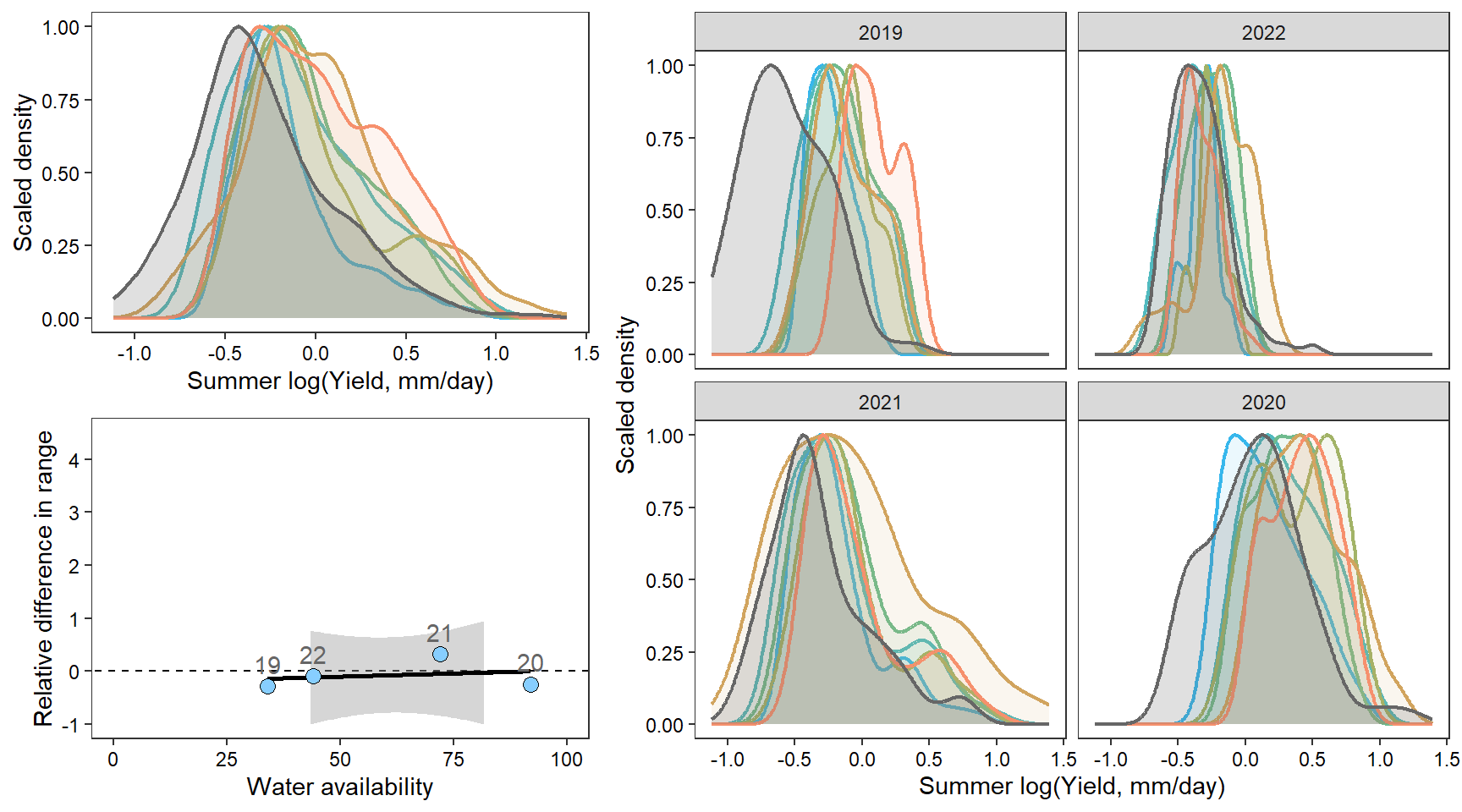
denfl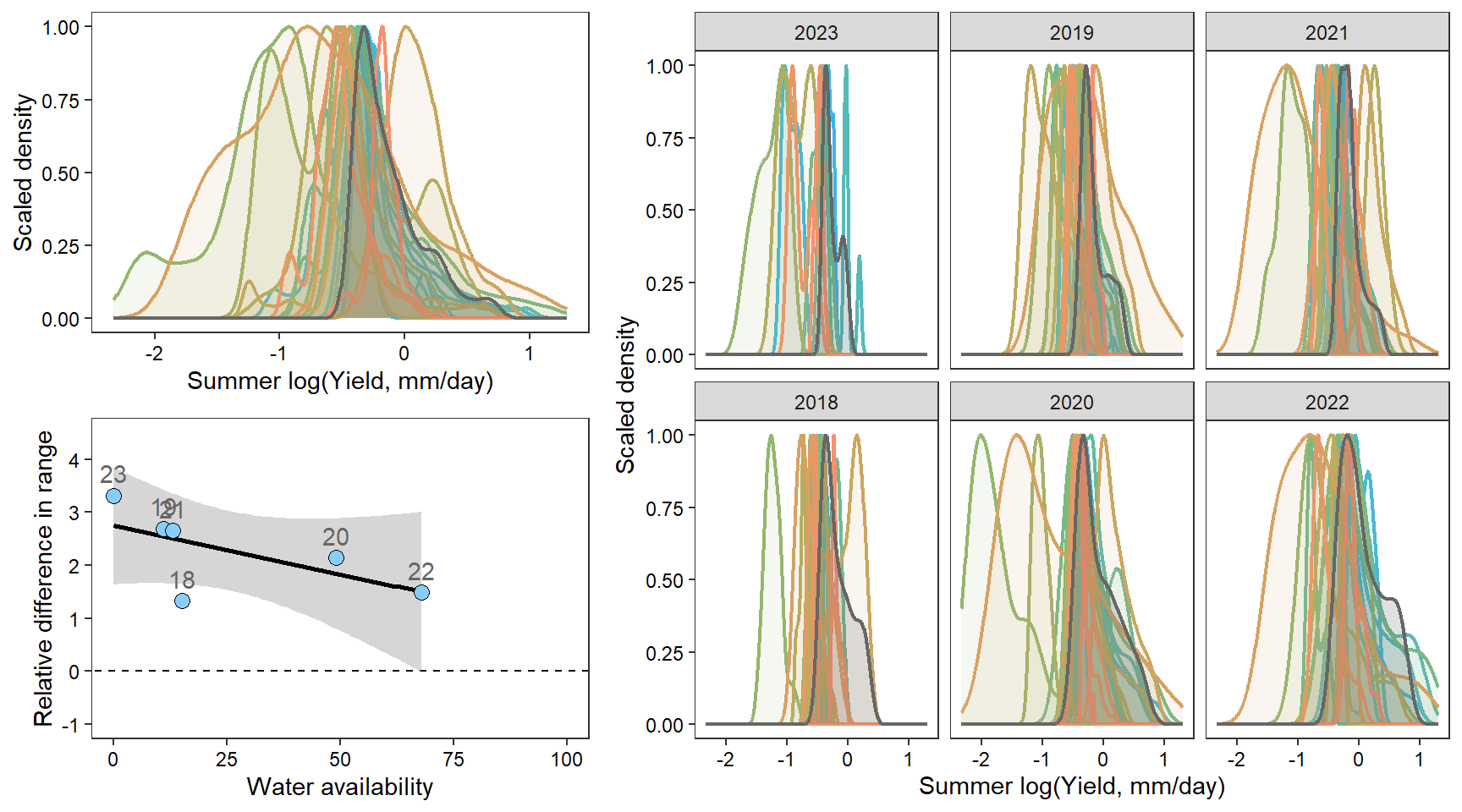
denye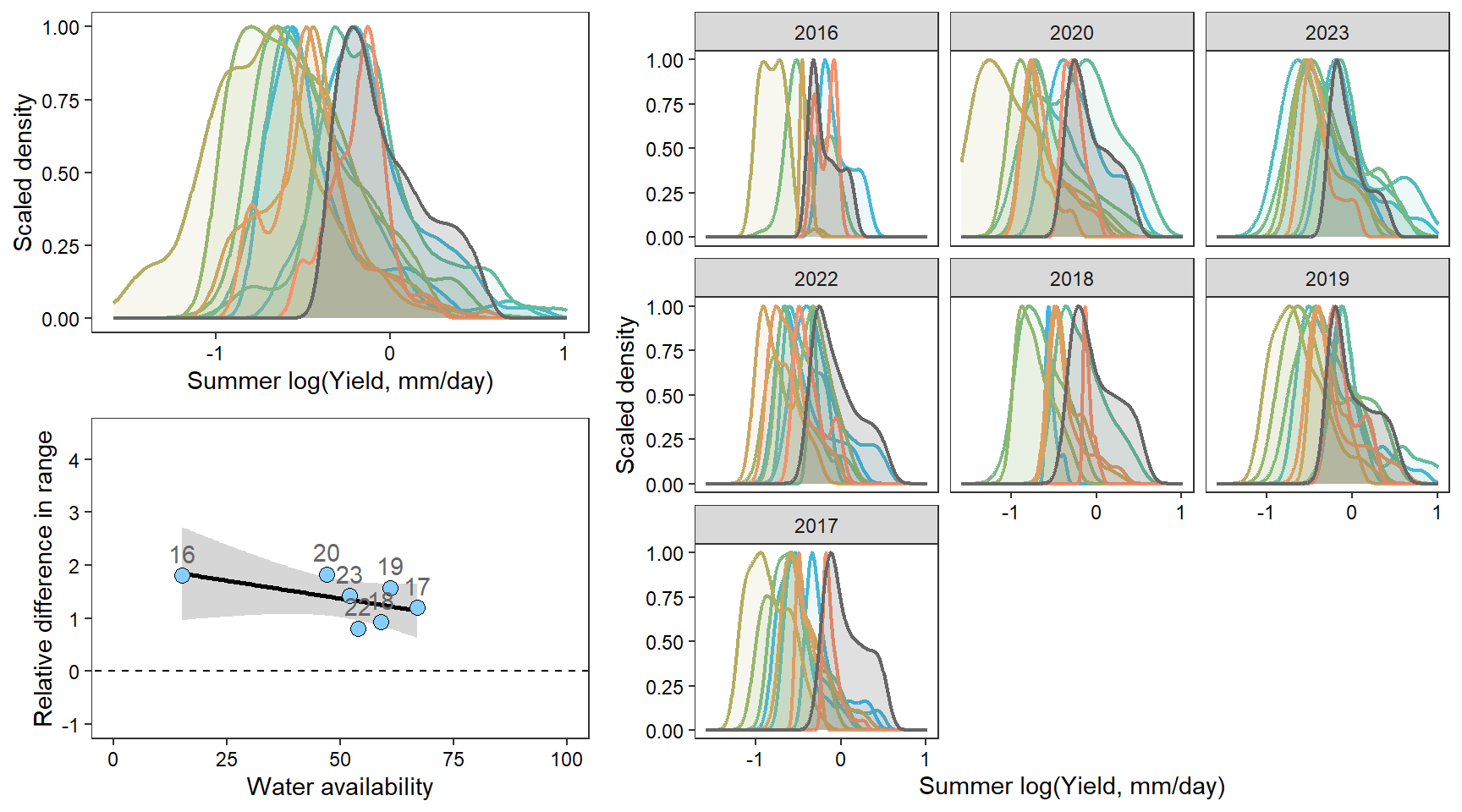
densn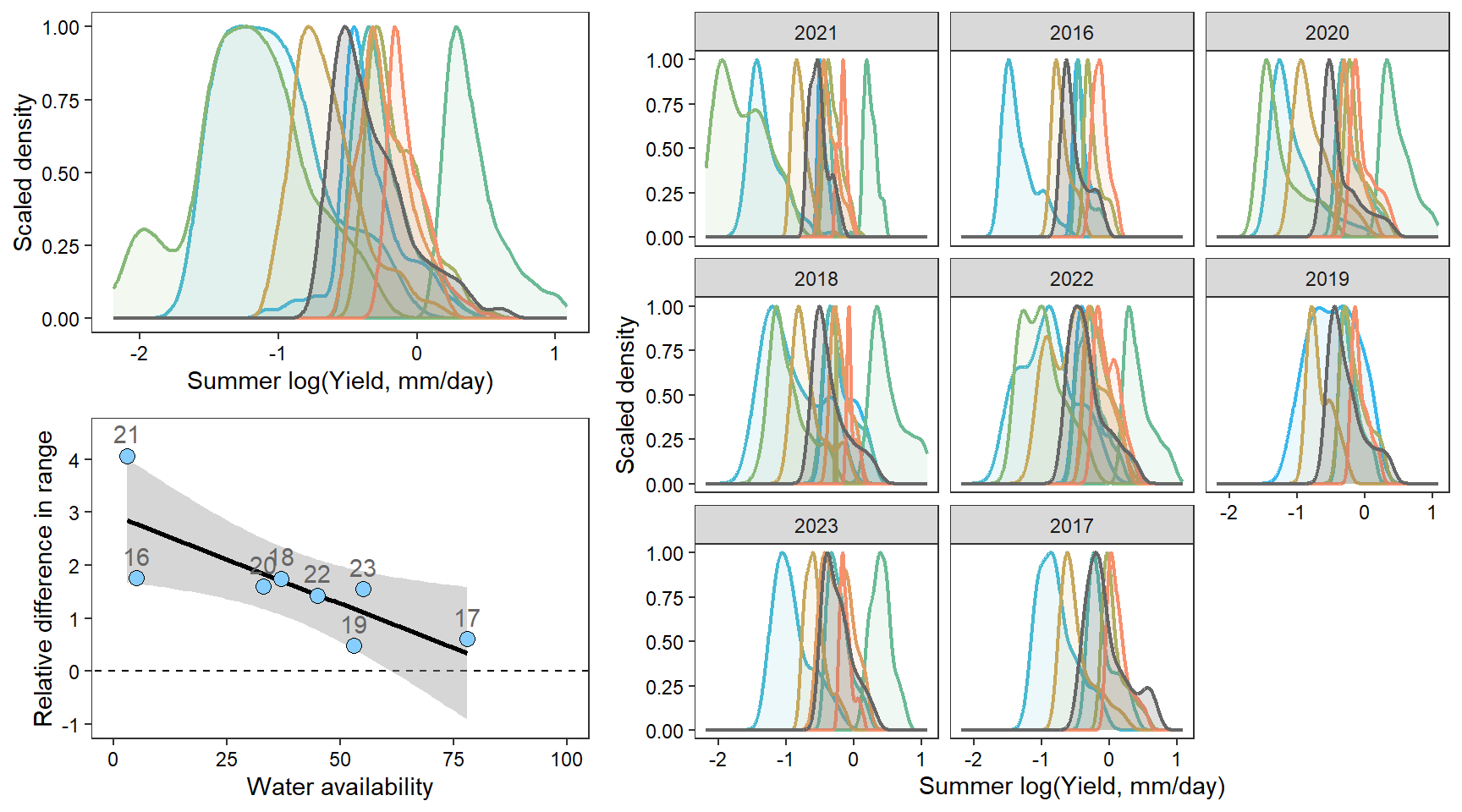
dendb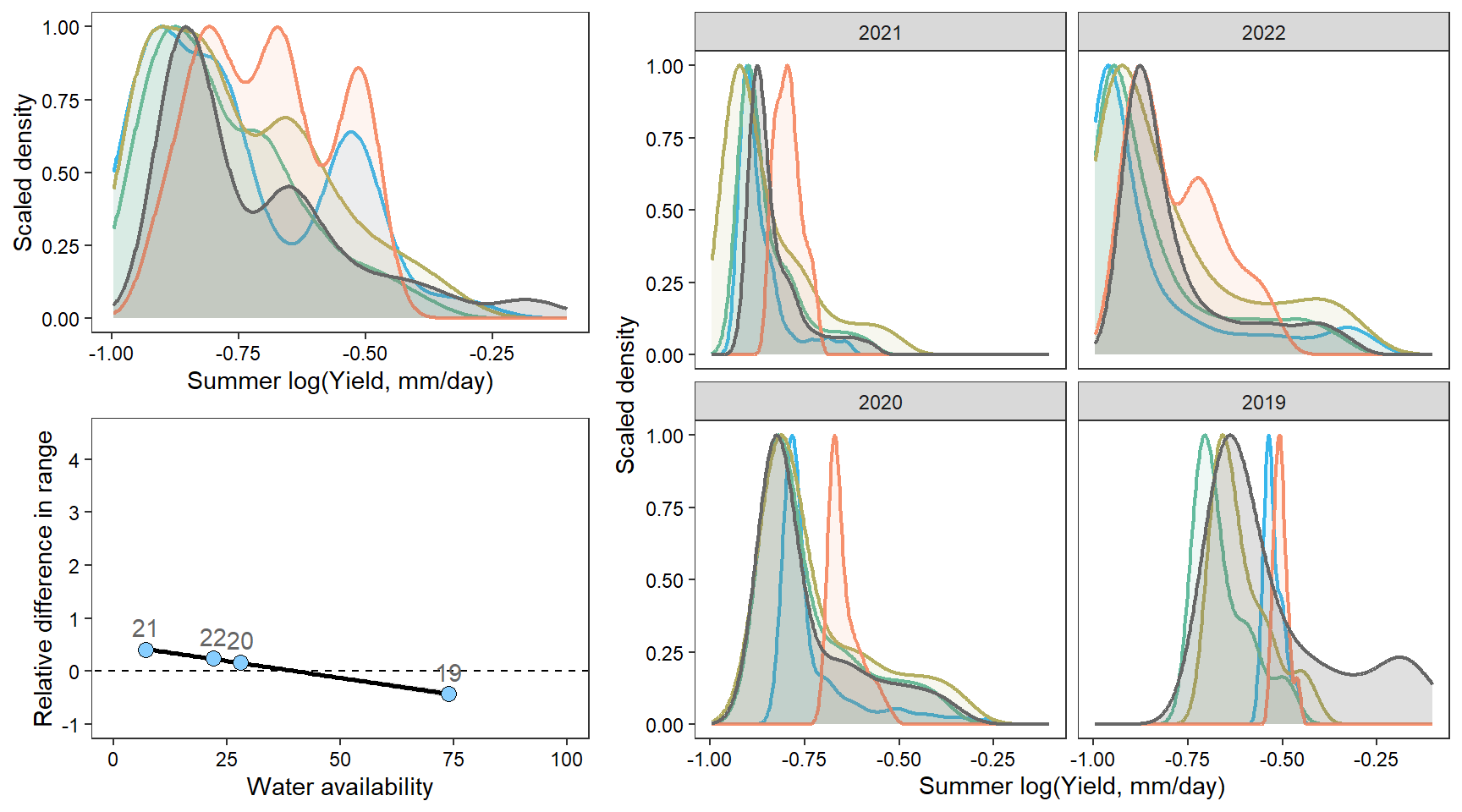
Plot interanntual summer log(yield) distributions for all basins
p <- egg::ggarrange(
# West Brook
portfolioplot(bas = "West Brook", orderr = wborder, type = "interannual", wtryrs = c(2020:2024)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 1, b = 2, l = 1), panel.border = element_rect(colour = mycols[1], fill = NA, linewidth = 1.5)),
# Staunton
portfolioplot(bas = "Staunton River", orderr = stauntorder, type = "interannual", wtryrs = c(2019:2022)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 1, b = 2, l = 1), panel.border = element_rect(colour = mycols[2], fill = NA, linewidth = 1.5)),
# Paine
portfolioplot(bas = "Paine Run", orderr = paineorder, type = "interannual", wtryrs = c(2019:2022)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 1, b = 2, l = 1), panel.border = element_rect(colour = mycols[3], fill = NA, linewidth = 1.5)),
# Flathead
portfolioplot(bas = "Flathead", orderr = flatorder, type = "interannual", wtryrs = c(2018:2024)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 1, b = 2, l = 1), panel.border = element_rect(colour = mycols[4], fill = NA, linewidth = 1.5)),
# Yellowstone
portfolioplot(bas = "Shields River", orderr = yellorder, type = "interannual", wtryrs = c(2016:2020,2022:2024)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 1, b = 2, l = 1), panel.border = element_rect(colour = mycols[5], fill = NA, linewidth = 1.5)),
# Snake
portfolioplot(bas = "Snake River", orderr = snakeorder, type = "interannual", wtryrs = c(2016:2024)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 1, b = 2, l = 1), panel.border = element_rect(colour = mycols[6], fill = NA, linewidth = 1.5)),
# Donner Blitzen
portfolioplot(bas = "Donner Blitzen", orderr = donnerorder, type = "interannual", wtryrs = c(2016:2024)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 1, b = 2, l = 1), panel.border = element_rect(colour = mycols[7], fill = NA, linewidth = 1.5)),
nrow = 7)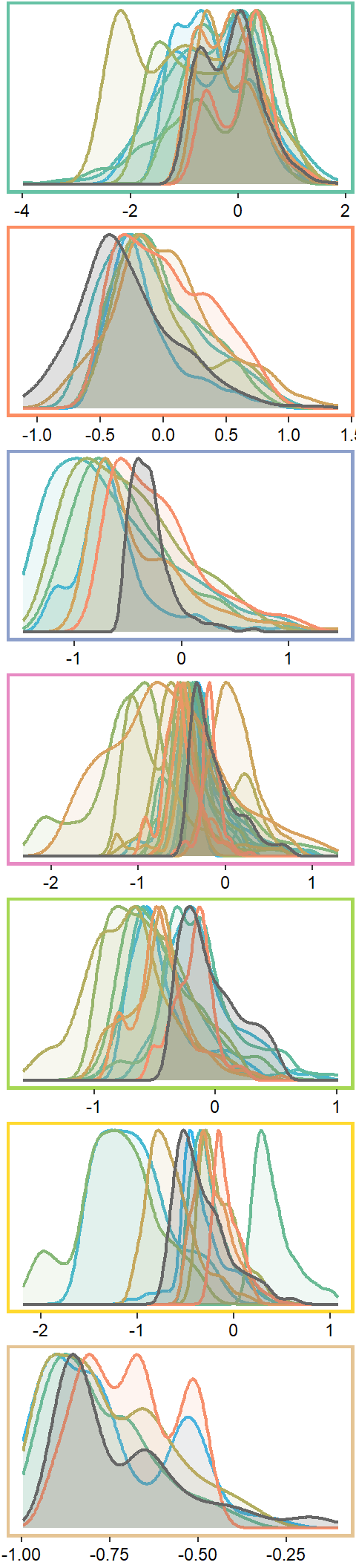
# no borders
p1 <- egg::ggarrange(
# West Brook
portfolioplot(bas = "West Brook", orderr = wborder, type = "interannual", wtryrs = c(2020:2024)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 7, b = 2, l = 1)),
# Staunton
portfolioplot(bas = "Staunton River", orderr = stauntorder, type = "interannual", wtryrs = c(2019:2022)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 7, b = 2, l = 1)),
# Paine
portfolioplot(bas = "Paine Run", orderr = paineorder, type = "interannual", wtryrs = c(2019:2022)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 7, b = 2, l = 1)),
# Flathead
portfolioplot(bas = "Flathead", orderr = flatorder, type = "interannual", wtryrs = c(2018:2024)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 7, b = 2, l = 1)),
# Yellowstone
portfolioplot(bas = "Shields River", orderr = yellorder, type = "interannual", wtryrs = c(2016:2020,2022:2024)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 7, b = 2, l = 1)),
# Snake
portfolioplot(bas = "Snake River", orderr = snakeorder, type = "interannual", wtryrs = c(2016:2024)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 7, b = 2, l = 1)),
# Donner Blitzen
portfolioplot(bas = "Donner Blitzen", orderr = donnerorder, type = "interannual", wtryrs = c(2016:2024)) +
theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(),
plot.margin = margin(t = 1, r = 7, b = 2, l = 1)),
nrow = 7)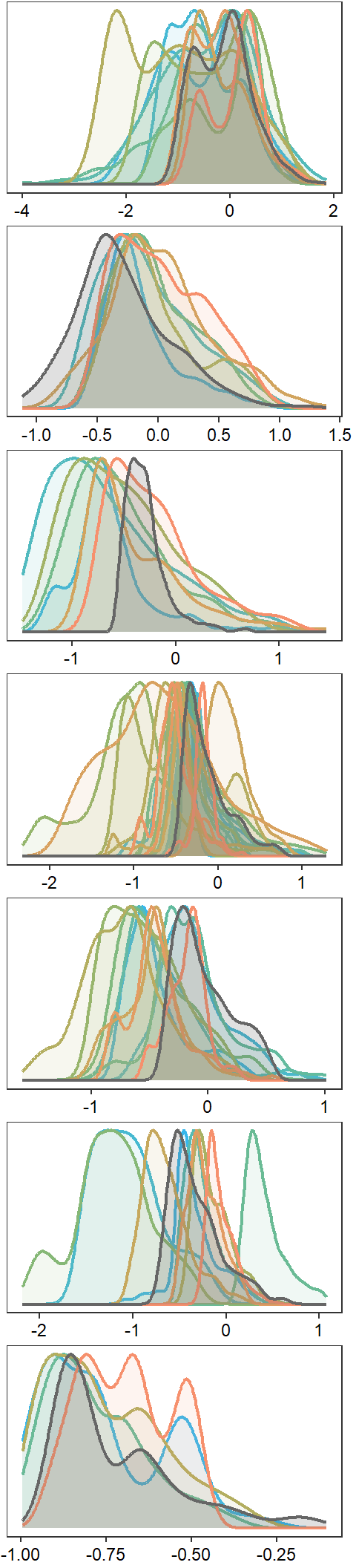
#annotate_figure(p, left = "Density")
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_summerdensity_all_noborder_wide.jpg", width = 2.5, height = 11, units = "in", res = 1000)
annotate_figure(p1, left = "Density")
dev.off()png
2 How does regional water availability affect relative difference range of streamflow values (headwaters vs. reference gages)?
Some data manipulation
# define basin-years to keep
keepyears <- bind_rows(
tibble(basin = "West Brook", WaterYear = c(2020:2024)),
tibble(basin = "Paine Run", WaterYear = c(2019:2022)),
tibble(basin = "Staunton River", WaterYear = c(2019:2022)),
tibble(basin = "Flathead", WaterYear = c(2018:2024)),
tibble(basin = "Shields River", WaterYear = c(2016:2020,2022:2024)),
tibble(basin = "Snake River", WaterYear = c(2016:2024)),
tibble(basin = "Donner Blitzen", WaterYear = c(2016:2024)),
) %>% mutate(keep = "yes")
# standard deviation of median summer flow
# dat_clean %>%
# filter(Month %in% c(7:9)) %>%
# group_by(basin, site_name, WaterYear) %>%
# summarize(medyield = median(logYield, na.rm = TRUE)) %>%
# ungroup() %>%
# group_by(basin, WaterYear) %>%
# summarize(sdyield = sd(medyield), nsites = n()) %>%
# ungroup() %>%
# left_join(wateravail %>% select(basin, WaterYear, totalyield:tyz_sum_perc)) %>%
# left_join(keepyears) %>% filter(keep == "yes") %>%
# mutate(basin = ifelse(basin == "Shields River", "Yellowstone River",
# ifelse(basin == "Flathead", "Flathead River", basin))) %>%
# mutate(basin = factor(basin, levels = c("West Brook", "Piney River", "Staunton River", "Paine Run", "Flathead River", "Yellowstone River", "Snake River", "Donner Blitzen"))) %>%
# ggplot(aes(x = tyz_sum_perc, y = sdyield)) +
# geom_smooth(method = "lm", color = "black", aes(weight = nsites)) +
# geom_point(aes(fill = basin, size = nsites), shape = 21) +
# scale_fill_brewer(palette = "Set2") +
# #facet_wrap(~basin) +
# theme_bw() + theme(panel.grid = element_blank())
# calculate relative difference in range as above
mydat <- dat_clean %>%
filter(Month %in% c(7:9)) %>%
group_by(basin, WaterYear) %>%
summarize(nsites = length(unique(site_name)),
littlerange = range(logYield)[2]-range(logYield)[1]) %>%
ungroup() %>%
left_join(dat_clean_big %>%
filter(Month %in% c(7:9)) %>%
group_by(basin, WaterYear) %>%
summarize(bigrange = range(logYield)[2]-range(logYield)[1]) %>%
ungroup()) %>%
mutate(pe_range = ((littlerange-bigrange)/bigrange)*100) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield:tyz_sum_perc)) %>%
left_join(keepyears) %>% filter(keep == "yes") %>%
mutate(basin = ifelse(basin == "Shields River", "Yellowstone River",
ifelse(basin == "Flathead", "Flathead River",
ifelse(basin == "Donner Blitzen", "Donner-Blitzen River", basin)))) %>%
mutate(basin = factor(basin, levels = c("West Brook", "Staunton River", "Paine Run",
"Flathead River", "Yellowstone River", "Snake River", "Donner-Blitzen River")))
mydat# A tibble: 38 × 14
basin WaterYear nsites littlerange bigrange pe_range site_name totalyield
<fct> <dbl> <int> <dbl> <dbl> <dbl> <chr> <dbl>
1 Donner-B… 2019 4 0.332 0.583 -43.1 Donner B… 278.
2 Donner-B… 2020 4 0.605 0.516 17.3 Donner B… 152.
3 Donner-B… 2021 4 0.474 0.336 41.1 Donner B… 122.
4 Donner-B… 2022 4 0.741 0.597 24.0 Donner B… 125.
5 Flathead… 2018 18 1.82 0.783 133. North Fo… 654.
6 Flathead… 2019 19 2.47 0.668 269. North Fo… 448.
7 Flathead… 2020 19 3.49 1.11 214. North Fo… 605.
8 Flathead… 2021 17 2.76 0.756 265. North Fo… 483.
9 Flathead… 2022 16 2.76 1.11 149. North Fo… 793.
10 Flathead… 2023 9 1.94 0.451 331. North Fo… 372.
# ℹ 28 more rows
# ℹ 6 more variables: totalyield_z <dbl>, totalyield_sum <dbl>,
# totalyield_sum_z <dbl>, tyz_perc <dbl>, tyz_sum_perc <dbl>, keep <chr>Show simple linear model summary, weighted by number of sites
summary(lm(pe_range ~ tyz_sum_perc, data = mydat, weights = nsites))
Call:
lm(formula = pe_range ~ tyz_sum_perc, data = mydat, weights = nsites)
Weighted Residuals:
Min 1Q Median 3Q Max
-503.62 -205.22 -25.09 108.84 639.45
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 239.1231 26.7291 8.946 1.12e-10 ***
tyz_sum_perc -1.8174 0.5199 -3.496 0.00127 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 262.6 on 36 degrees of freedom
Multiple R-squared: 0.2534, Adjusted R-squared: 0.2327
F-statistic: 12.22 on 1 and 36 DF, p-value: 0.001273Show distribution of spatial streamflow heterogeneity by basin. Lots of structuring by basin, driven in large part by Staunton and Donner-Blitzen Rivers.
par(mar = c(9,4,0.5,0.5))
boxplot(pe_range ~ basin, data = mydat, ylab = "Spatial streamflow heterogeneity", xlab = "", las = 2, col = mycols)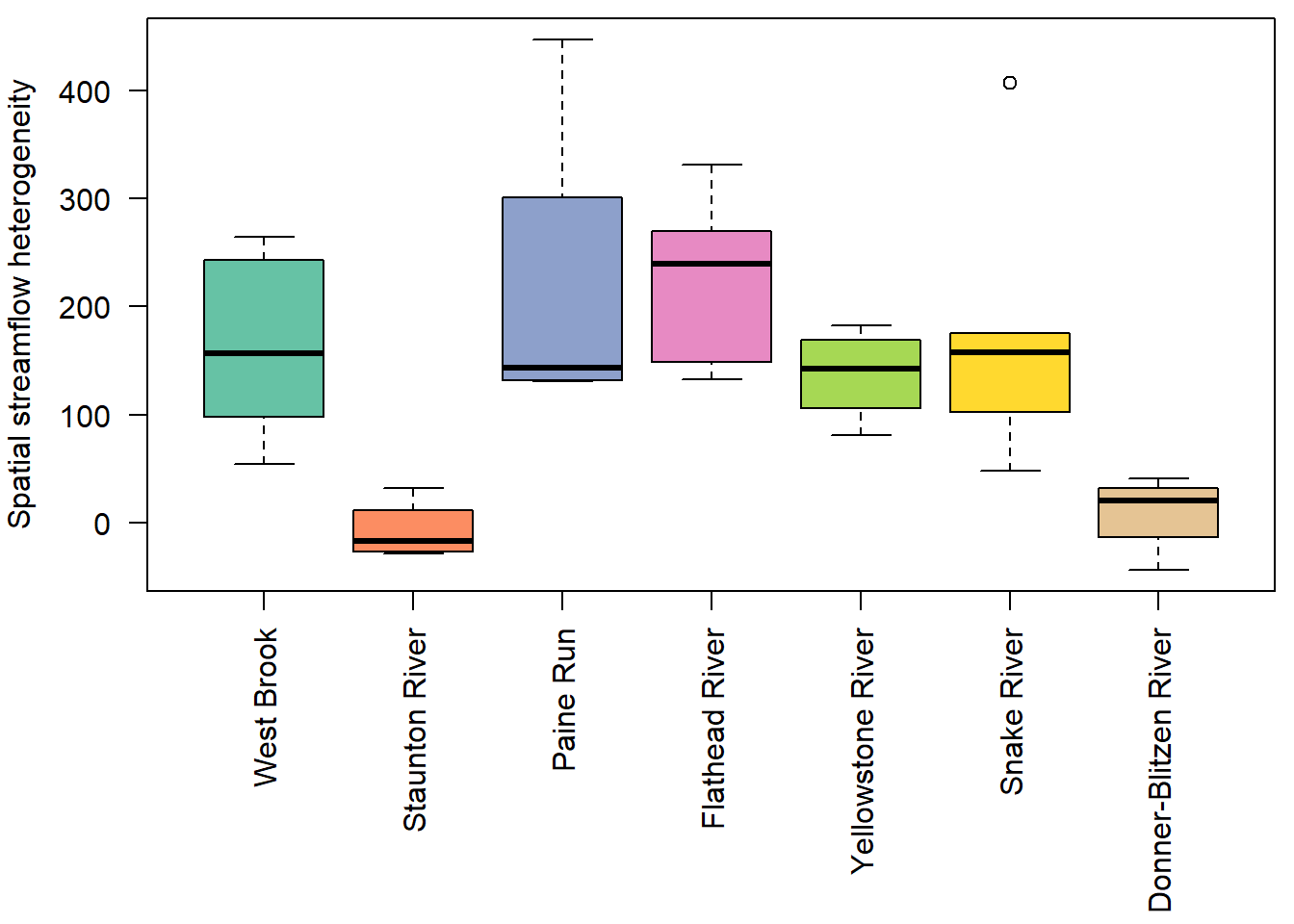
Use a random slope/random intercept mixed effects model to estimate the among-basin mean effect of regional water availability on spatial streamflow heterogeneity.
Notes:
mydat <- mydat %>% mutate(z_tyz_sum_perc = scale(tyz_sum_perc))
lmod <- lmer(pe_range ~ z_tyz_sum_perc + (1 + z_tyz_sum_perc | basin), data = mydat)
summary(lmod)Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: pe_range ~ z_tyz_sum_perc + (1 + z_tyz_sum_perc | basin)
Data: mydat
REML criterion at convergence: 430.4
Scaled residuals:
Min 1Q Median 3Q Max
-1.5512 -0.4206 -0.1253 0.2584 2.8102
Random effects:
Groups Name Variance Std.Dev. Corr
basin (Intercept) 7810.1 88.37
z_tyz_sum_perc 609.8 24.69 -0.92
Residual 5127.9 71.61
Number of obs: 38, groups: basin, 7
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 133.13 35.60 5.51 3.740 0.0113 *
z_tyz_sum_perc -45.39 16.29 3.62 -2.787 0.0554 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr)
z_tyz_sm_pr -0.521# r.squaredGLMM(lmod)Get p-values for global slope and intercept: (doesn’t to run in Quarto…works fine in R)
coef(summary(as_lmerModLmerTest(lmod))) Estimate Std. Error df t value Pr(>|t|)
(Intercept) 133.13376 35.60169 5.510014 3.739534 0.01125212
z_tyz_sum_perc -45.39251 16.28830 3.620345 -2.786817 0.05538425Plot relationship between spatial streamflow heterogeneity and regional water availability with gloabl (among-population mean) regression line.
# get predicted values and 95% credible interval
effects_tyz <- effects::effect(term = "z_tyz_sum_perc", mod = as_lmerModLmerTest(lmod), xlevels = list(z_tyz_sum_perc = seq(min(mydat$z_tyz_sum_perc), max(mydat$z_tyz_sum_perc), length.out = 1000)), se = list(level = 0.95))
x_tyz <- as_tibble(effects_tyz) %>% mutate(tyz_sum_perc = (z_tyz_sum_perc * attr(mydat$z_tyz_sum_perc, "scaled:scale")) + attr(mydat$z_tyz_sum_perc, "scaled:center"))
# trim data for panel labesl
mydat_labs <- bind_rows(mydat %>% filter(basin == "West Brook", WaterYear %in% c(2020,2021)),
mydat %>% filter(basin == "Snake River", WaterYear %in% c(2021,2022))) %>%
mutate(labs = c("b", "c", "d", "e"))
# plot
randiffplot <- ggplot() +
geom_ribbon(data = x_tyz, aes(x = tyz_sum_perc, ymin = lower, ymax = upper), fill = "grey80") +
geom_line(data = x_tyz, aes(x = tyz_sum_perc, y = fit), color = "black", size = 1.25, lineend = "round") +
geom_point(data = mydat, aes(x = tyz_sum_perc, y = pe_range, fill = basin, size = nsites), shape = 21) +
scale_fill_brewer(palette = "Set2") +
geom_text(data = mydat_labs, aes(x = tyz_sum_perc, y = pe_range, label = labs), size = 3) +
theme_bw() + theme(panel.grid = element_blank(), axis.text = element_text(color = "black")) +
ylab("Spatial streamflow heterogeneity (percent difference)") + xlab("Water availability (percentile)") +
guides(fill = guide_legend(title = "Basin", override.aes = list(size = 5), order = 1), size = guide_legend(title = "Number of sites", order = 2)) +
geom_abline(intercept = 0, slope = 0, linetype = "dashed")
randiffplot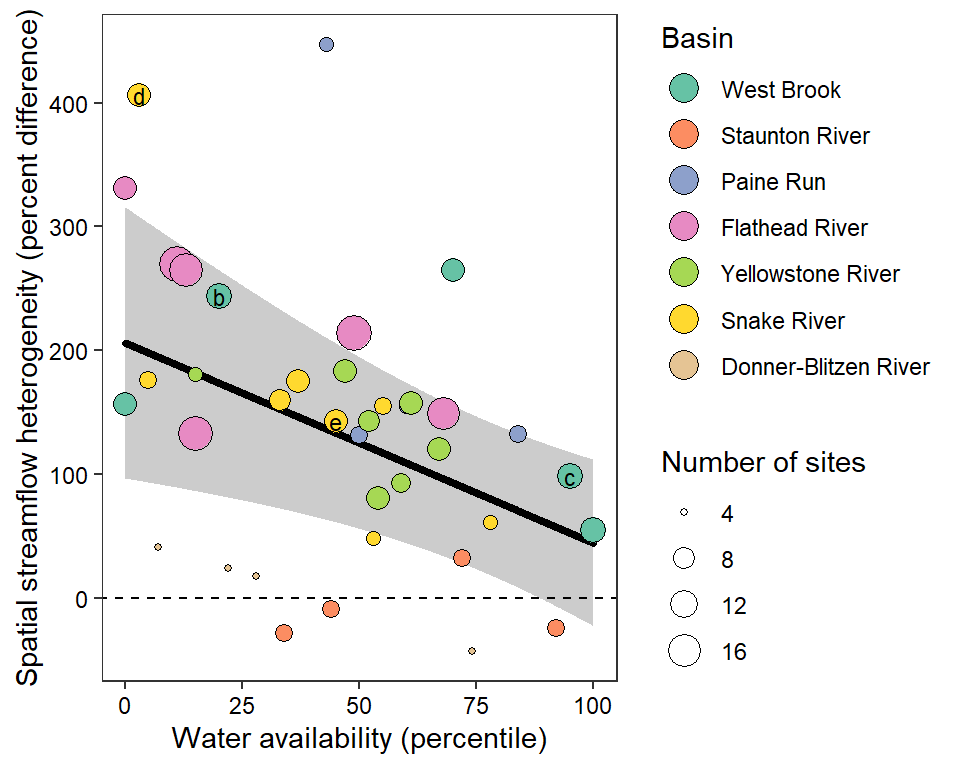
# write to file
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_RangeDiff_allsites2.jpg", width = 4.75, height = 5, units = "in", res = 1000)
randiffplot
dev.off()png
2 Use rptR to decompose variance in spatial streamflow heterogeneity into fixed (regional water availability) and random (basin) effect sources:
rmod <- rptGaussian(pe_range ~ z_tyz_sum_perc + (1 + z_tyz_sum_perc | basin), data = mydat, grname = c("basin", "Fixed"), ratio = TRUE, adjusted = FALSE, nboot = 1000)Bootstrap Progress:print(rmod)
Repeatability estimation using the lmm method
Repeatability for basin
R = 0.539
SE = 0.169
CI = [0.154, 0.801]
P = 0.00764 [LRT]
NA [Permutation]
Repeatability for Fixed
R = 0.132
SE = 0.099
CI = [0.012, 0.378]
P = NA [LRT]
NA [Permutation]rmod$modLinear mixed model fit by REML ['lmerMod']
Formula: pe_range ~ z_tyz_sum_perc + (1 + z_tyz_sum_perc | basin)
Data: data
REML criterion at convergence: 430.4229
Random effects:
Groups Name Std.Dev. Corr
basin (Intercept) 88.37
z_tyz_sum_perc 24.69 -0.92
Residual 71.61
Number of obs: 38, groups: basin, 7
Fixed Effects:
(Intercept) z_tyz_sum_perc
133.13 -45.39 plot(rmod, grname = "basin")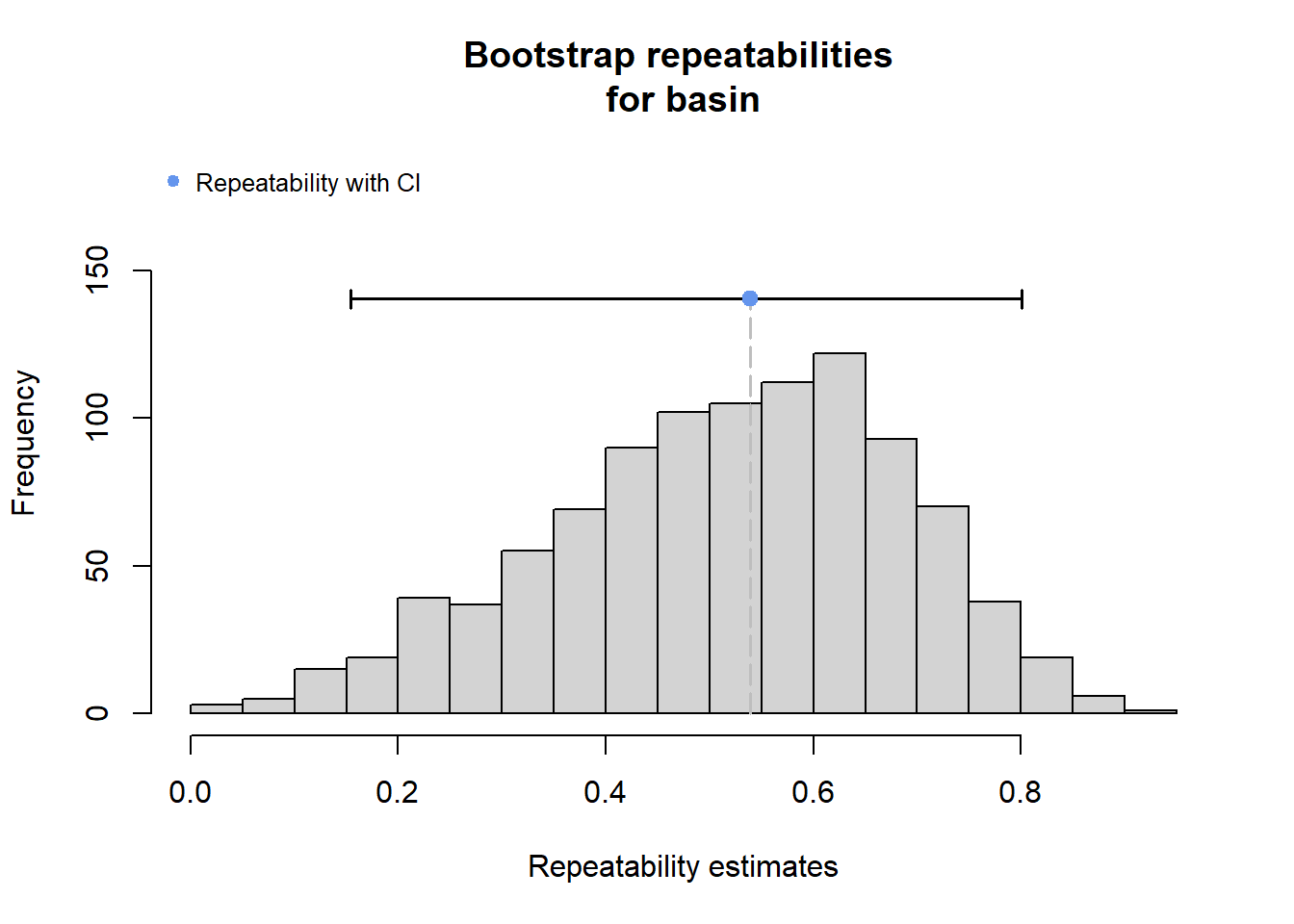
plot(rmod, grname = "Fixed")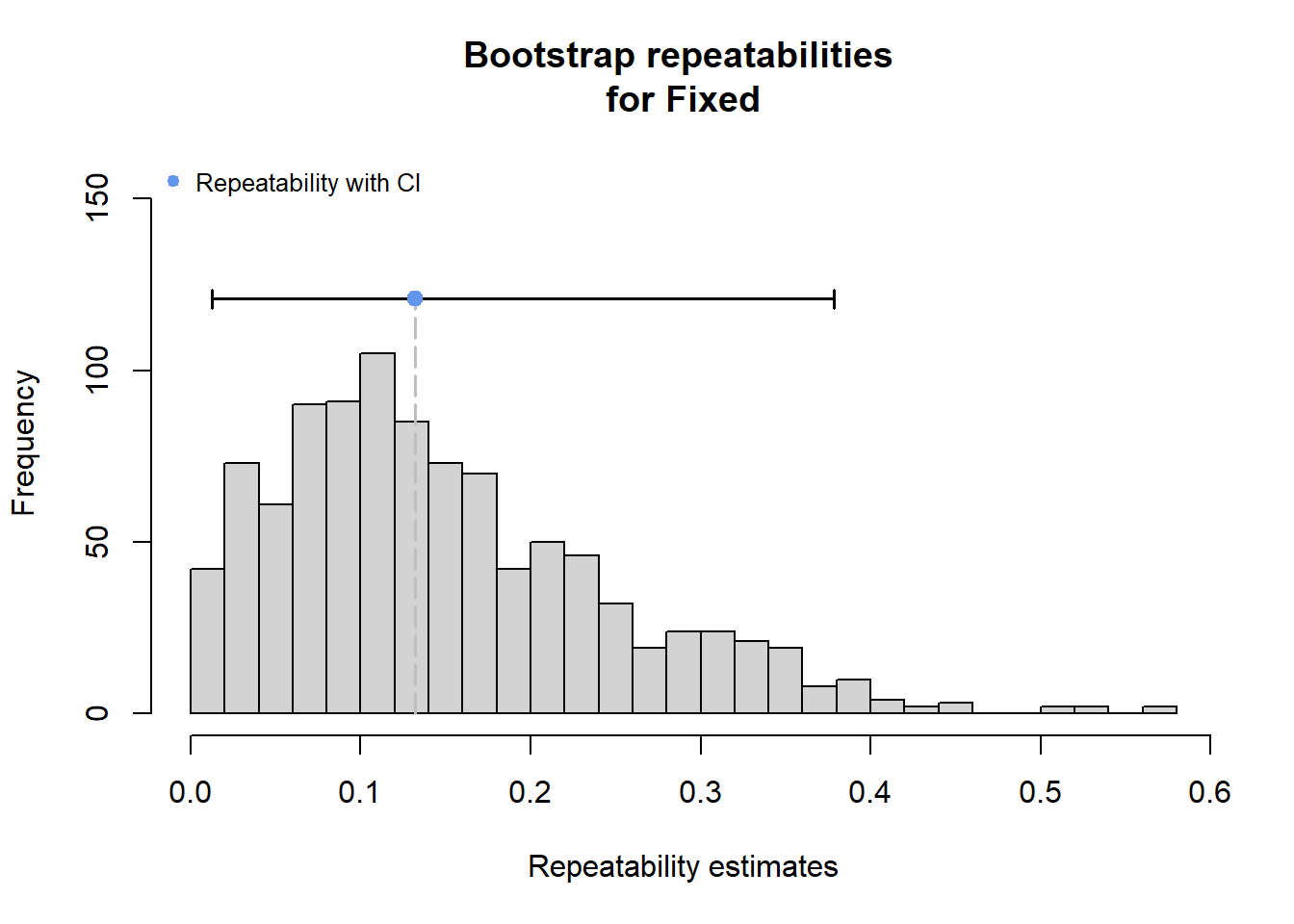
The strong effect of basin is largely driven by Staunton and Donner-Blitzen Rivers. Without those basins, random effect variance shrinks and fixed effect variance expands considerably. Much of this pattern is likely due to low sample sizes (few headwater gages in these basins, particularly in the Donner-Blitzen)
rmod <- rptGaussian(pe_range ~ z_tyz_sum_perc + (1 + z_tyz_sum_perc | basin), data = mydat %>% filter(!basin %in% c("Staunton River", "Donner-Blitzen River")), grname = c("basin", "Fixed"), ratio = TRUE, adjusted = FALSE, nboot = 1000)Bootstrap Progress:print(rmod)
Repeatability estimation using the lmm method
Repeatability for basin
R = 0.037
SE = 0.11
CI = [0, 0.392]
P = 1 [LRT]
NA [Permutation]
Repeatability for Fixed
R = 0.304
SE = 0.132
CI = [0.062, 0.574]
P = NA [LRT]
NA [Permutation]How sensitive are these results to the choice of data (log) transformation, sensu comment from John Hammond during internal review?
Summary: While the among-network mean effect of water availability on heterogeneity was less significant using un-logged discharge data (p > 0.05), this appeared to be driven by the highly non-normal distribution of raw discharge values, as the reviewer indicated. Importantly, the magnitude of the effect is similar and the qualitative results are unchanged (negative slope).
Data manipulation: use un-logged/raw Yield data to calculation spatial heterogeneity, also use CV rather than ranges
mydat2 <- dat_clean %>%
filter(Month %in% c(7:9)) %>%
group_by(basin, WaterYear) %>%
summarize(nsites = length(unique(site_name)),
littlerange = range(Yield_mm)[2]-range(Yield_mm)[1],
littlecv = sd(Yield_mm)/mean(Yield_mm)) %>%
ungroup() %>%
left_join(dat_clean_big %>%
filter(Month %in% c(7:9)) %>%
group_by(basin, WaterYear) %>%
summarize(bigrange = range(Yield_mm)[2]-range(Yield_mm)[1],
bigcv = sd(Yield_mm)/mean(Yield_mm)) %>%
ungroup()) %>%
mutate(pe_range = ((littlerange-bigrange)/bigrange)*100,
pe_cv = ((littlecv-bigcv)/bigcv)*100) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield:tyz_sum_perc)) %>%
left_join(keepyears) %>% filter(keep == "yes") %>%
mutate(basin = ifelse(basin == "Shields River", "Yellowstone River",
ifelse(basin == "Flathead", "Flathead River",
ifelse(basin == "Donner Blitzen", "Donner-Blitzen River", basin)))) %>%
mutate(basin = factor(basin, levels = c("West Brook", "Staunton River", "Paine Run",
"Flathead River", "Yellowstone River", "Snake River", "Donner-Blitzen River"))) Fig the mmixed effects model
mydat2 <- mydat2 %>% mutate(z_tyz_sum_perc = scale(tyz_sum_perc))
lmod <- lmer(pe_range ~ z_tyz_sum_perc + (1 + z_tyz_sum_perc | basin), data = mydat2)
summary(lmod)Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: pe_range ~ z_tyz_sum_perc + (1 + z_tyz_sum_perc | basin)
Data: mydat2
REML criterion at convergence: 523.5
Scaled residuals:
Min 1Q Median 3Q Max
-1.8092 -0.4147 -0.1069 0.3663 3.7590
Random effects:
Groups Name Variance Std.Dev. Corr
basin (Intercept) 79567 282.1
z_tyz_sum_perc 19495 139.6 -1.00
Residual 70913 266.3
Number of obs: 38, groups: basin, 7
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 238.131 116.273 4.215 2.048 0.106
z_tyz_sum_perc -86.826 72.670 2.761 -1.195 0.325
Correlation of Fixed Effects:
(Intr)
z_tyz_sm_pr -0.703
optimizer (nloptwrap) convergence code: 0 (OK)
boundary (singular) fit: see help('isSingular')Plot
# get predicted values and 95% credible interval
effects_tyz <- effects::effect(term = "z_tyz_sum_perc", mod = as_lmerModLmerTest(lmod), xlevels = list(z_tyz_sum_perc = seq(min(mydat2$z_tyz_sum_perc), max(mydat2$z_tyz_sum_perc), length.out = 1000)), se = list(level = 0.95))
x_tyz <- as_tibble(effects_tyz) %>% mutate(tyz_sum_perc = (z_tyz_sum_perc * attr(mydat2$z_tyz_sum_perc, "scaled:scale")) + attr(mydat2$z_tyz_sum_perc, "scaled:center"))
# trim data for panel labesl
mydat_labs <- bind_rows(mydat2 %>% filter(basin == "West Brook", WaterYear %in% c(2020,2021)),
mydat2 %>% filter(basin == "Snake River", WaterYear %in% c(2021,2022))) %>%
mutate(labs = c("b", "c", "d", "e"))
# plot
ggplot() +
geom_ribbon(data = x_tyz, aes(x = tyz_sum_perc, ymin = lower, ymax = upper), fill = "grey80") +
geom_line(data = x_tyz, aes(x = tyz_sum_perc, y = fit), color = "black", size = 1.25, lineend = "round") +
geom_point(data = mydat2, aes(x = tyz_sum_perc, y = pe_range, fill = basin, size = nsites), shape = 21) +
scale_fill_brewer(palette = "Set2") +
theme_bw() + theme(panel.grid = element_blank(), axis.text = element_text(color = "black")) +
ylab("Spatial streamflow heterogeneity (percent difference)") + xlab("Water availability (percentile)") +
guides(fill = guide_legend(title = "Basin", override.aes = list(size = 5), order = 1), size = guide_legend(title = "Number of sites", order = 2)) +
geom_abline(intercept = 0, slope = 0, linetype = "dashed")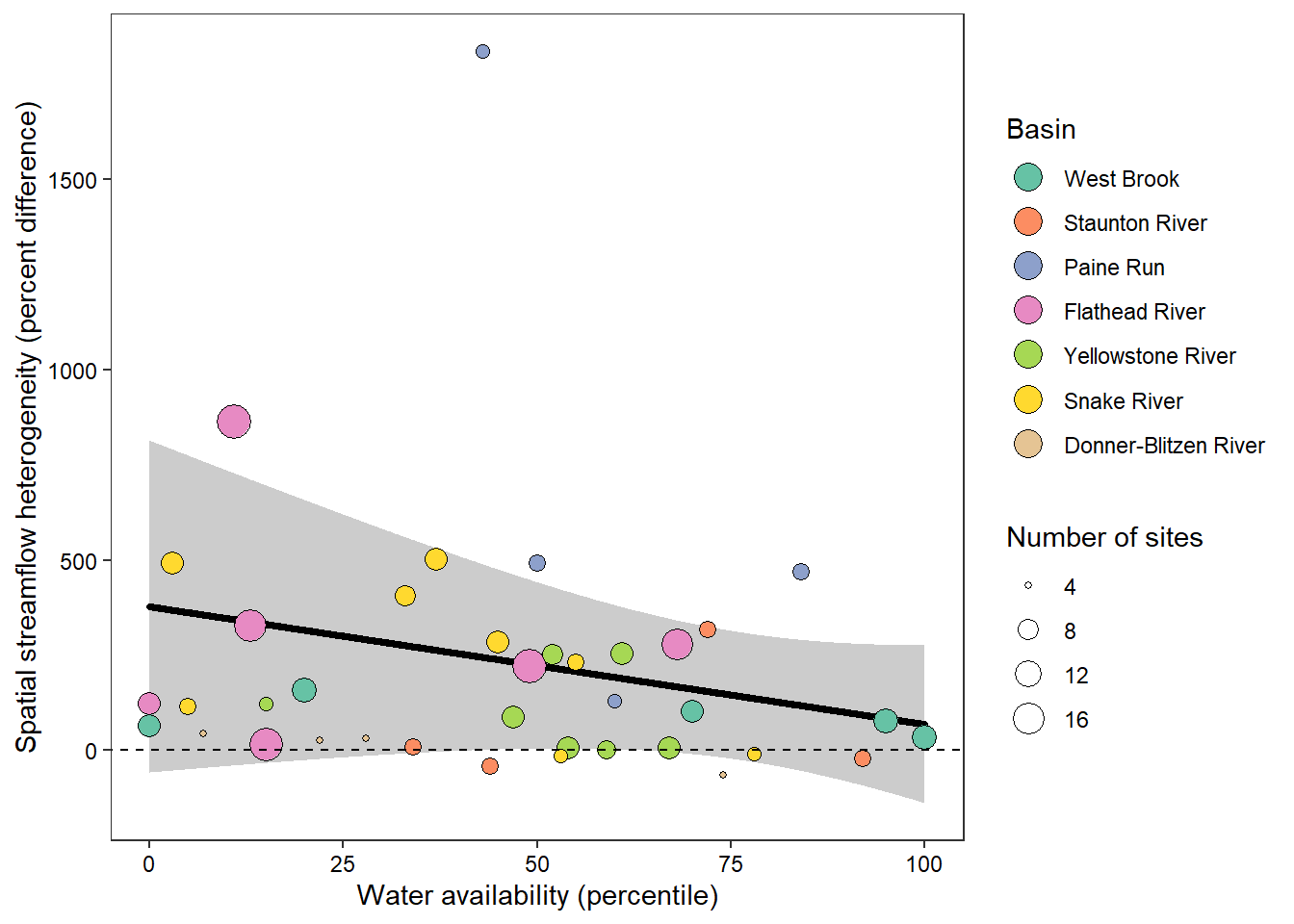
Fig the mmixed effects model
mydat2 <- mydat2 %>% mutate(z_tyz_sum_perc = scale(tyz_sum_perc))
lmod <- lmer(pe_cv ~ z_tyz_sum_perc + (1 + z_tyz_sum_perc | basin), data = mydat2)
summary(lmod)Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: pe_cv ~ z_tyz_sum_perc + (1 + z_tyz_sum_perc | basin)
Data: mydat2
REML criterion at convergence: 450.7
Scaled residuals:
Min 1Q Median 3Q Max
-2.4210 -0.3359 0.0006 0.2983 3.9794
Random effects:
Groups Name Variance Std.Dev. Corr
basin (Intercept) 24752 157.33
z_tyz_sum_perc 8948 94.59 -1.00
Residual 7991 89.39
Number of obs: 38, groups: basin, 7
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 88.405 61.551 5.248 1.436 0.208
z_tyz_sum_perc -55.189 39.739 3.675 -1.389 0.243
Correlation of Fixed Effects:
(Intr)
z_tyz_sm_pr -0.889
optimizer (nloptwrap) convergence code: 0 (OK)
boundary (singular) fit: see help('isSingular')Plot
# get predicted values and 95% credible interval
effects_tyz <- effects::effect(term = "z_tyz_sum_perc", mod = as_lmerModLmerTest(lmod), xlevels = list(z_tyz_sum_perc = seq(min(mydat2$z_tyz_sum_perc), max(mydat2$z_tyz_sum_perc), length.out = 1000)), se = list(level = 0.95))
x_tyz <- as_tibble(effects_tyz) %>% mutate(tyz_sum_perc = (z_tyz_sum_perc * attr(mydat2$z_tyz_sum_perc, "scaled:scale")) + attr(mydat2$z_tyz_sum_perc, "scaled:center"))
# trim data for panel labesl
mydat_labs <- bind_rows(mydat2 %>% filter(basin == "West Brook", WaterYear %in% c(2020,2021)),
mydat2 %>% filter(basin == "Snake River", WaterYear %in% c(2021,2022))) %>%
mutate(labs = c("b", "c", "d", "e"))
# plot
ggplot() +
geom_ribbon(data = x_tyz, aes(x = tyz_sum_perc, ymin = lower, ymax = upper), fill = "grey80") +
geom_line(data = x_tyz, aes(x = tyz_sum_perc, y = fit), color = "black", size = 1.25, lineend = "round") +
geom_point(data = mydat2, aes(x = tyz_sum_perc, y = pe_cv, fill = basin, size = nsites), shape = 21) +
scale_fill_brewer(palette = "Set2") +
theme_bw() + theme(panel.grid = element_blank(), axis.text = element_text(color = "black")) +
ylab("Spatial streamflow heterogeneity (percent difference)") + xlab("Water availability (percentile)") +
guides(fill = guide_legend(title = "Basin", override.aes = list(size = 5), order = 1), size = guide_legend(title = "Number of sites", order = 2)) +
geom_abline(intercept = 0, slope = 0, linetype = "dashed")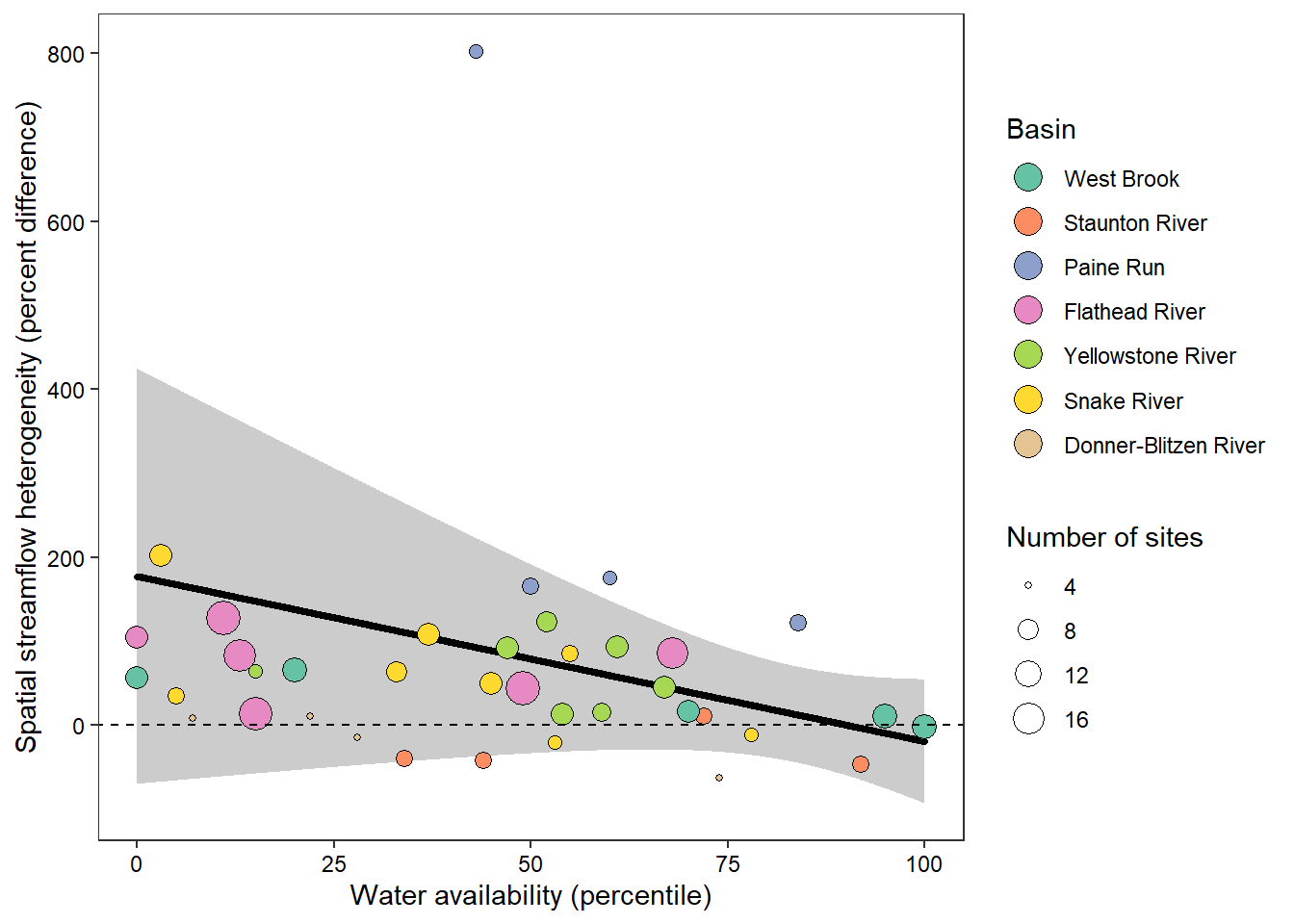
bas <- "West Brook"
orderr <- wborder
wtryrs <- c(2020:2024)
# sheds and network
mysheds <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/Mass_Watersheds.shp")
mysheds <- mysheds[mysheds$site_id == "WBR",]
mynet <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams/Mass_Streams.shp")
crs(mynet) <- crs(mysheds)
mynet <- crop(mynet, mysheds)
# get lakes
lakes <- get_waterbodies(AOI = siteinfo_sp %>% filter(site_name == "West Brook NWIS"), buffer = 10000)
lakes <- lakes %>% filter(gnis_name %in% c("Northampton Reservoir Upper", "Northampton Reservoir"))
#st_write(lakes, "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Study area maps/lakes/lakes_WestBrook.shp")
# little g points
mylittleg <- siteinfo_sp %>% filter(site_name %in% wborder) %>% mutate(site_name = factor(site_name, levels = wborder))
# filter data
tempdat <- dat_clean %>%
filter(basin == bas, !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = orderr))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == bas, !is.na(logYield), Month %in% c(7:9))
# calculate variability for reference gage
varbig <- tempdat_big %>%
group_by(WaterYear) %>%
summarize(bigrange = range(logYield)[2]-range(logYield)[1],
bigsd = sd(logYield),
bigvar = var(logYield)) %>%
ungroup()
# calculate total summer water availability from reference gage
#summerflow <- tempdat_big %>% group_by(WaterYear) %>% summarize(summeryield = sum(Yield_mm), summeryield_log = log(sum(Yield_mm))) %>% ungroup()
#wateravail2 <- wateravail %>% select(basin, WaterYear, totalyield, totalyield_z)
# calculate relative variation
pedat <- tempdat %>%
group_by(basin, WaterYear) %>%
summarize(nsites = length(unique(site_name)),
littlerange = range(logYield)[2]-range(logYield)[1],
littlesd = sd(logYield),
littlevar = var(logYield)) %>%
ungroup() %>%
left_join(varbig) %>%
mutate(pe_range = ((littlerange-bigrange)/bigrange),
pe_sd = ((littlesd-bigsd)/bigsd),
pe_var = ((littlevar-bigvar)/bigvar)) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield, totalyield_z, totalyield_sum, totalyield_sum_z, tyz_perc, tyz_sum_perc)) %>%
filter(WaterYear %in% wtryrs) %>%
mutate(wylab = substr(WaterYear, 3, 4)) %>%
arrange(totalyield_sum_z)
# calculate annual median summer flow
annmed <- tempdat %>%
#filter(WaterYear %in% c(lowyr, highyr)) %>%
group_by(site_name, WaterYear) %>%
summarize(medlogYield = median(logYield, na.rm = TRUE)) %>%
ungroup()
# calculate long-term median of summer median flow, to center color scheme
annmed_big <- tempdat_big %>%
group_by(site_name, WaterYear) %>%
summarize(medlogYield = median(logYield, na.rm = TRUE)) %>%
ungroup()
annmed_big <- median(annmed_big$medlogYield)
# arrange years by total annual yield
wtryrs_arranged <- as.numeric(unlist(pedat %>% select(WaterYear)))
# plot it
plotlist_wb <- list()
for(i in 1:length(wtryrs_arranged)) {
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i]))
mylittleg_sheds <- sheds %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i])) %>% arrange(desc(area_sqmi))
plotlist_wb[[i]] <- local({
i <- i
ggplot() +
geom_sf(data = st_as_sf(mysheds), color = "black", fill = "white", linewidth = 0.4) +
geom_sf(data = mylittleg_sheds, aes(fill = medlogYield), color = "black") +
scale_fill_viridis(option = "G", direction = 1, limits = range(annmed$medlogYield), na.value = "grey") +
#scale_fill_gradientn(colours = rev(hcl.colors(100, "Zissou1")), limits = range(annmed$medlogYield), na.value = "grey") +
#scale_fill_gradientn(colours = cet_pal(100, name = "l16"), limits = range(annmed$medlogYield), na.value = "grey") +
geom_sf(data = st_as_sf(mynet), color = "white", linewidth = 1, lineend = "round") +
geom_sf(data = st_as_sf(mynet), color = "royalblue4", linewidth = 0.6, lineend = "round") +
geom_sf(data = lakes, color = "white", fill = "lightskyblue1", linewidth = 0.7) +
geom_sf(data = lakes, color = "royalblue4", fill = "lightskyblue1", linewidth = 0.5) +
geom_sf(data = mylittleg, shape = 21, fill = "white", size = 2) +
labs(fill = "Median\nsummer\nlog(Yield)") + #annotation_scale() +
coord_sf(xlim = range(st_coordinates(mylittleg_sheds)[,1]), ylim = range(st_coordinates(mylittleg_sheds)[,2])) +
theme_bw() + theme(axis.title.x = element_blank(), axis.title.y = element_blank(), legend.position = "none", axis.text = element_blank()) +
annotate("label", x = Inf, y = Inf, label = paste(pedat$WaterYear[i], " (", pedat$tyz_sum_perc[i], "%)", sep = ""), vjust = 1.15, hjust = 1.05, size = 5)})
}
ggarrange(plotlist = plotlist_wb, common.legend = TRUE, legend = "right")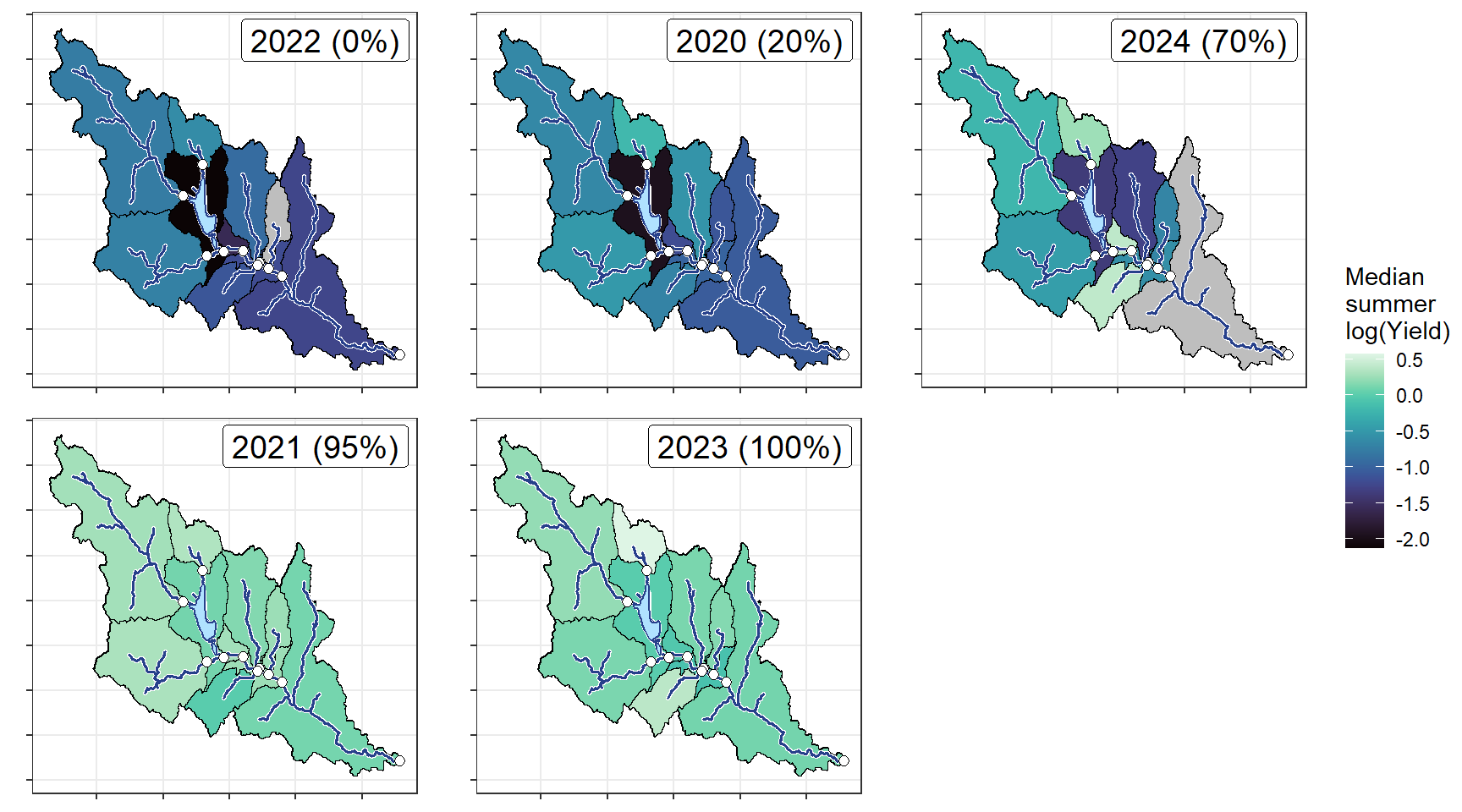
bas <- "Paine Run"
orderr <- paineorder
wtryrs <- c(2019:2022)
mysheds <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/Shen_Watersheds.shp")
mysheds <- mysheds[mysheds$site_id == "PA_10FL",]
mynet <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams/Shen_Streams.shp")
crs(mynet) <- crs(mysheds)
mynet <- crop(mynet, mysheds)
# hillshade
myrast <- rast("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Data/Spatial data/Elevation/Shenandoah_DEM_10m_nc.tif")
myrast <- mask(crop(myrast, mysheds), mysheds)
slo <- terrain(myrast, "slope", unit = "radians")
asp <- terrain(myrast, "aspect", unit = "radians")
hill <- shade(slope = slo, aspect = asp, angle = 40, direction = 270)
hilldf <- as.data.frame(hill, xy = TRUE)
# little g points
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr)
# filter data
tempdat <- dat_clean %>%
filter(basin == bas, !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = orderr))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% unique(tempdat$WaterYear))
# calculate variability for reference gage
varbig <- tempdat_big %>%
group_by(WaterYear) %>%
summarize(bigrange = range(logYield)[2]-range(logYield)[1],
bigsd = sd(logYield),
bigvar = var(logYield)) %>%
ungroup()
# calculate total summer water availability from reference gage
#summerflow <- tempdat_big %>% group_by(WaterYear) %>% summarize(summeryield = sum(Yield_mm), summeryield_log = log(sum(Yield_mm))) %>% ungroup()
#wateravail2 <- wateravail %>% select(basin, WaterYear, totalyield, totalyield_z)
# calculate relative variation
pedat <- tempdat %>%
group_by(basin, WaterYear) %>%
summarize(nsites = length(unique(site_name)),
littlerange = range(logYield)[2]-range(logYield)[1],
littlesd = sd(logYield),
littlevar = var(logYield)) %>%
ungroup() %>%
left_join(varbig) %>%
mutate(pe_range = ((littlerange-bigrange)/bigrange),
pe_sd = ((littlesd-bigsd)/bigsd),
pe_var = ((littlevar-bigvar)/bigvar)) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield, totalyield_z, totalyield_sum, totalyield_sum_z, tyz_perc, tyz_sum_perc)) %>%
filter(WaterYear %in% wtryrs) %>%
mutate(wylab = substr(WaterYear, 3, 4)) %>%
arrange(totalyield_sum_z)
# calculate annual median summer flow
annmed <- tempdat %>%
#filter(WaterYear %in% c(lowyr, highyr)) %>%
group_by(site_name, WaterYear) %>%
summarize(medlogYield = median(logYield, na.rm = TRUE)) %>%
ungroup()
# arrange years by total annual yield
wtryrs_arranged <- as.numeric(unlist(pedat %>% select(WaterYear)))
# plot it
plotlist_pa <- list()
for(i in 1:length(wtryrs_arranged)) {
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i]))
mylittleg_sheds <- sheds %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i])) %>% arrange(desc(area_sqmi))
plotlist_pa[[i]] <- local({
i <- i
ggplot() +
geom_sf(data = st_as_sf(mysheds), color = "black", fill = "white", linewidth = 0.4) +
geom_sf(data = mylittleg_sheds, aes(fill = medlogYield), color = "black") +
scale_fill_viridis(option = "G", direction = 1, limits = range(annmed$medlogYield), na.value = "grey") +
geom_sf(data = st_as_sf(mynet), color = "white", linewidth = 1, lineend = "round") +
geom_sf(data = st_as_sf(mynet), color = "royalblue4", linewidth = 0.6, lineend = "round") +
geom_sf(data = mylittleg, shape = 21, fill = "white", size = 2) +
labs(fill = "Median\nsummer\nlog(Yield)") + #annotation_scale() +
coord_sf(xlim = range(st_coordinates(mylittleg_sheds)[,1]), ylim = range(st_coordinates(mylittleg_sheds)[,2])) +
theme_bw() + theme(axis.title.x = element_blank(), axis.title.y = element_blank(), legend.position = "none", axis.text = element_blank()) +
annotate("label", x = -Inf, y = Inf, label = paste(pedat$WaterYear[i], " (", pedat$tyz_sum_perc[i], "%)", sep = ""), vjust = 1.15, hjust = -0.05, size = 5)})
}
ggarrange(plotlist = plotlist_pa, common.legend = TRUE, legend = "right")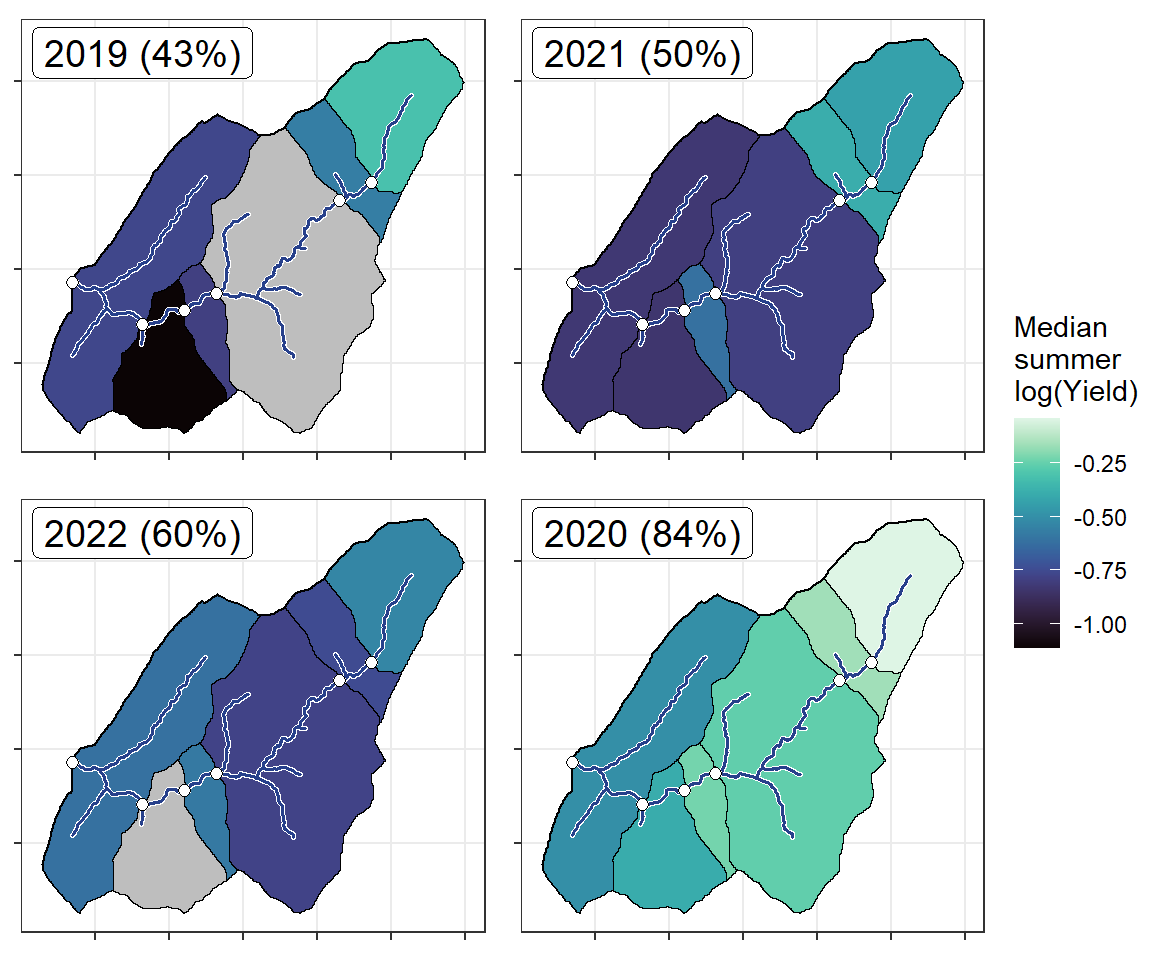
bas <- "Staunton River"
orderr <- stauntorder
wtryrs <- c(2019:2022)
mysheds <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/Shen_Watersheds.shp")
mysheds <- mysheds[mysheds$site_id == "SR_10FL",]
mynet <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams/Shen_Streams.shp")
crs(mynet) <- crs(mysheds)
mynet <- crop(mynet, mysheds)
# little g points
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr)
# filter data
tempdat <- dat_clean %>%
filter(basin == bas, !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = orderr))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% unique(tempdat$WaterYear))
# calculate variability for reference gage
varbig <- tempdat_big %>%
group_by(WaterYear) %>%
summarize(bigrange = range(logYield)[2]-range(logYield)[1],
bigsd = sd(logYield),
bigvar = var(logYield)) %>%
ungroup()
# calculate total summer water availability from reference gage
#summerflow <- tempdat_big %>% group_by(WaterYear) %>% summarize(summeryield = sum(Yield_mm), summeryield_log = log(sum(Yield_mm))) %>% ungroup()
#wateravail2 <- wateravail %>% select(basin, WaterYear, totalyield, totalyield_z)
# calculate relative variation
pedat <- tempdat %>%
group_by(basin, WaterYear) %>%
summarize(nsites = length(unique(site_name)),
littlerange = range(logYield)[2]-range(logYield)[1],
littlesd = sd(logYield),
littlevar = var(logYield)) %>%
ungroup() %>%
left_join(varbig) %>%
mutate(pe_range = ((littlerange-bigrange)/bigrange),
pe_sd = ((littlesd-bigsd)/bigsd),
pe_var = ((littlevar-bigvar)/bigvar)) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield, totalyield_z, totalyield_sum, totalyield_sum_z, tyz_perc, tyz_sum_perc)) %>%
filter(WaterYear %in% wtryrs) %>%
mutate(wylab = substr(WaterYear, 3, 4)) %>%
arrange(totalyield_sum_z)
# calculate annual median summer flow
annmed <- tempdat %>%
#filter(WaterYear %in% c(lowyr, highyr)) %>%
group_by(site_name, WaterYear) %>%
summarize(medlogYield = median(logYield, na.rm = TRUE)) %>%
ungroup()
# arrange years by total annual yield
wtryrs_arranged <- as.numeric(unlist(pedat %>% select(WaterYear)))
# plot it
plotlist_st <- list()
for(i in 1:length(wtryrs_arranged)) {
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i]))
mylittleg_sheds <- sheds %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i])) %>% arrange(desc(area_sqmi))
plotlist_st[[i]] <- local({
i <- i
ggplot() +
geom_sf(data = st_as_sf(mysheds), color = "black", fill = "white", linewidth = 0.4) +
geom_sf(data = mylittleg_sheds, aes(fill = medlogYield), color = "black") +
scale_fill_viridis(option = "G", direction = 1, limits = range(annmed$medlogYield), na.value = "grey") +
geom_sf(data = st_as_sf(mynet), color = "white", linewidth = 1, lineend = "round") +
geom_sf(data = st_as_sf(mynet), color = "royalblue4", linewidth = 0.6, lineend = "round") +
geom_sf(data = mylittleg, shape = 21, fill = "white", size = 2) +
labs(fill = "Median\nsummer\nlog(Yield)") + #annotation_scale() +
coord_sf(xlim = range(st_coordinates(mylittleg_sheds)[,1]), ylim = range(st_coordinates(mylittleg_sheds)[,2])) +
theme_bw() + theme(axis.title.x = element_blank(), axis.title.y = element_blank(), legend.position = "none", axis.text = element_blank()) +
annotate("label", x = Inf, y = Inf, label = paste(pedat$WaterYear[i], " (", pedat$tyz_sum_perc[i], "%)", sep = ""), vjust = 1.15, hjust = 1.05, size = 5)})
}
ggarrange(plotlist = plotlist_st, common.legend = TRUE, legend = "right")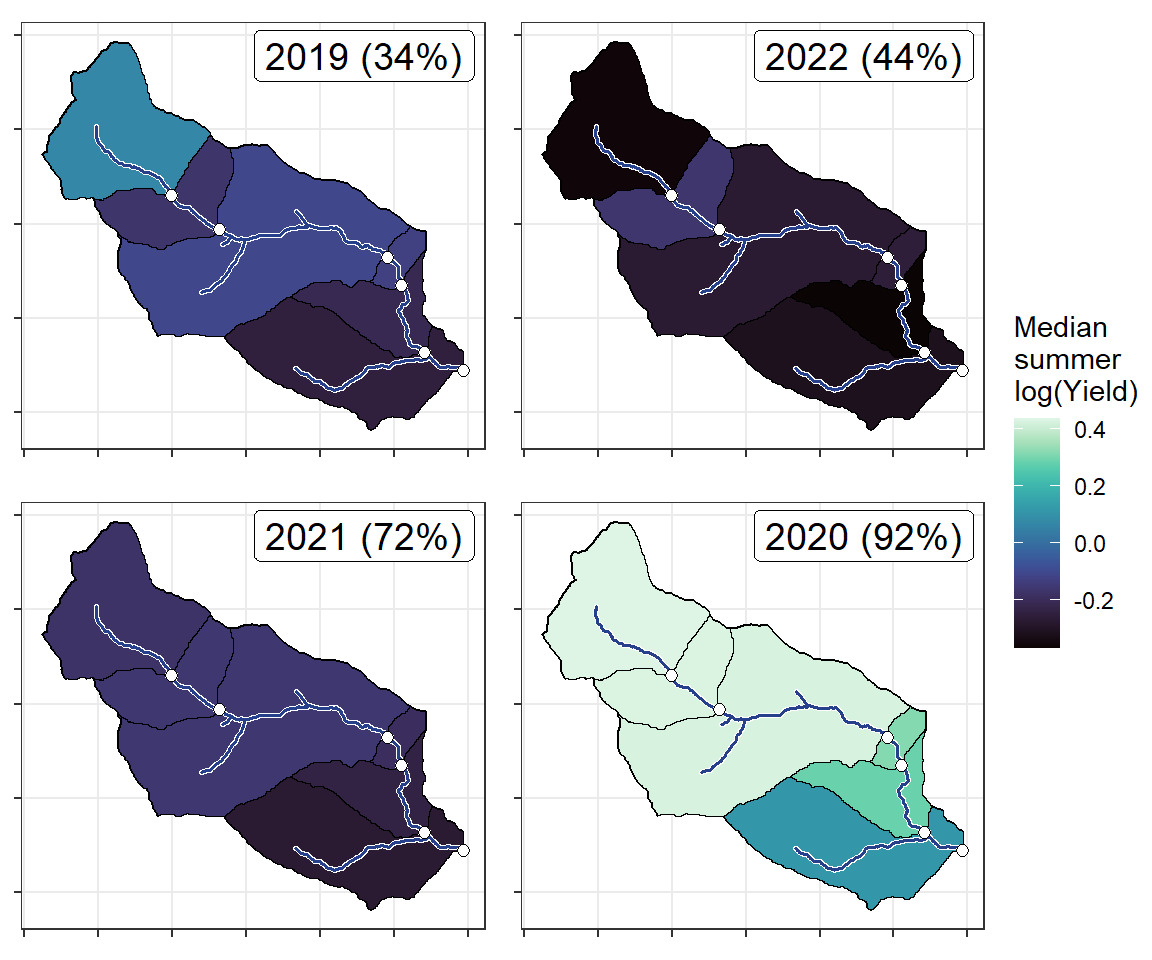
bas <- "Flathead"
orderr <- flatorder
wtryrs <- c(2018:2024)
mysheds <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/Flat_Watersheds.shp")
mysheds <- mysheds[mysheds$site_id == "NFF",]
mynet <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams/Flat_Streams.shp")
crs(mynet) <- crs(mysheds)
mynet <- crop(mynet, mysheds)
# get lakes
lakes <- get_waterbodies(AOI = siteinfo_sp %>% filter(site_name == "North Fork Flathead River NWIS"), buffer = 100000)
lakes <- st_transform(lakes, crs(mysheds))
lakes <- st_intersection(lakes, st_as_sf(mysheds))
lakes <- lakes %>% filter(gnis_name %in% c("Moose Lake", "Mud Lake"))
# points
mylittleg <- siteinfo_sp %>% filter(site_name %in% flatorder) %>% mutate(site_name = factor(site_name, levels = flatorder))
st_geometry(mylittleg)[mylittleg$site_name == "BigCreekUpper"] <- st_point(c(-114.31506, 48.57672))
st_geometry(mylittleg)[mylittleg$site_name == "HallowattCreekLower"] <- st_point(c(-114.31914, 48.57256))
# filter data
tempdat <- dat_clean %>%
filter(basin == bas, !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = orderr))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% unique(tempdat$WaterYear))
# calculate variability for reference gage
varbig <- tempdat_big %>%
group_by(WaterYear) %>%
summarize(bigrange = range(logYield)[2]-range(logYield)[1],
bigsd = sd(logYield),
bigvar = var(logYield)) %>%
ungroup()
# calculate total summer water availability from reference gage
#summerflow <- tempdat_big %>% group_by(WaterYear) %>% summarize(summeryield = sum(Yield_mm), summeryield_log = log(sum(Yield_mm))) %>% ungroup()
#wateravail2 <- wateravail %>% select(basin, WaterYear, totalyield, totalyield_z)
# calculate relative variation
pedat <- tempdat %>%
group_by(basin, WaterYear) %>%
summarize(nsites = length(unique(site_name)),
littlerange = range(logYield)[2]-range(logYield)[1],
littlesd = sd(logYield),
littlevar = var(logYield)) %>%
ungroup() %>%
left_join(varbig) %>%
mutate(pe_range = ((littlerange-bigrange)/bigrange),
pe_sd = ((littlesd-bigsd)/bigsd),
pe_var = ((littlevar-bigvar)/bigvar)) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield, totalyield_z, totalyield_sum, totalyield_sum_z, tyz_perc, tyz_sum_perc)) %>%
filter(WaterYear %in% wtryrs) %>%
mutate(wylab = substr(WaterYear, 3, 4)) %>%
arrange(totalyield_sum_z)
# calculate annual median summer flow
annmed <- tempdat %>%
#filter(WaterYear %in% c(lowyr, highyr)) %>%
group_by(site_name, WaterYear) %>%
summarize(medlogYield = median(logYield, na.rm = TRUE)) %>%
ungroup()
# arrange years by total annual yield
wtryrs_arranged <- as.numeric(unlist(pedat %>% select(WaterYear)))
# plot it
plotlist_fl <- list()
for(i in 1:length(wtryrs_arranged)) {
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i]))
mylittleg_sheds <- sheds %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i])) %>% arrange(desc(area_sqmi))
plotlist_fl[[i]] <- local({
i <- i
ggplot() +
geom_sf(data = st_as_sf(mysheds), color = "black", fill = "white", linewidth = 0.4) +
geom_sf(data = mylittleg_sheds, aes(fill = medlogYield), color = "black") +
scale_fill_viridis(option = "G", direction = 1, limits = range(annmed$medlogYield), na.value = "grey") +
geom_sf(data = st_as_sf(mynet), color = "white", linewidth = 1, lineend = "round") +
geom_sf(data = st_as_sf(mynet), color = "royalblue4", linewidth = 0.6, lineend = "round") +
geom_sf(data = mylittleg, shape = 21, fill = "white", size = 2) +
labs(fill = "Median\nsummer\nlog(Yield)") + #annotation_scale() +
coord_sf(xlim = range(st_coordinates(mylittleg_sheds)[,1]), ylim = range(st_coordinates(mylittleg_sheds)[,2])) +
theme_bw() + theme(axis.title.x = element_blank(), axis.title.y = element_blank(), legend.position = "none", axis.text = element_blank()) +
annotate("label", x = Inf, y = Inf, label = paste(pedat$WaterYear[i], " (", pedat$tyz_sum_perc[i], "%)", sep = ""), vjust = 1.15, hjust = 1.05, size = 5)})
}
ggarrange(plotlist = plotlist_fl, common.legend = TRUE, legend = "right")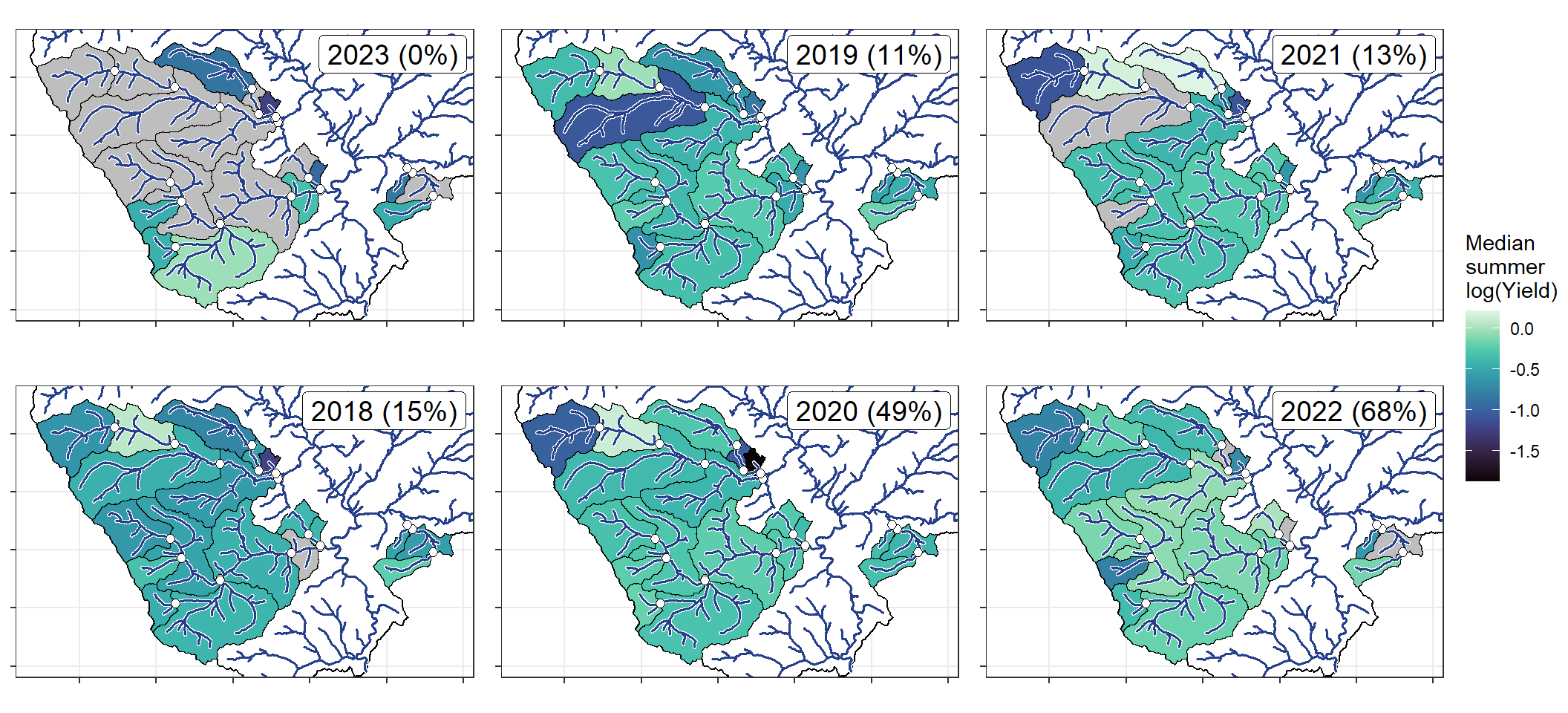
bas <- "Shields River"
orderr <- yellorder
wtryrs <- c(2016:2020,2022:2024)
mysheds <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/Shields_Watersheds.shp")
mysheds <- mysheds[mysheds$site_id %in% c("SRS", "DU01"),]
mynet <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams/Shields_Streams.shp")
crs(mynet) <- crs(mysheds)
mynet <- crop(mynet, mysheds)
# points
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr) %>% mutate(site_name = factor(site_name, levels = orderr)) #%>% filter(subbasin != "Duck Creek")
# filter data
tempdat <- dat_clean %>%
filter(basin == bas, !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = orderr))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% unique(tempdat$WaterYear))
# calculate variability for reference gage
varbig <- tempdat_big %>%
group_by(WaterYear) %>%
summarize(bigrange = range(logYield)[2]-range(logYield)[1],
bigsd = sd(logYield),
bigvar = var(logYield)) %>%
ungroup()
# calculate total summer water availability from reference gage
#summerflow <- tempdat_big %>% group_by(WaterYear) %>% summarize(summeryield = sum(Yield_mm), summeryield_log = log(sum(Yield_mm))) %>% ungroup()
#wateravail2 <- wateravail %>% select(basin, WaterYear, totalyield, totalyield_z)
# calculate relative variation
pedat <- tempdat %>%
group_by(basin, WaterYear) %>%
summarize(nsites = length(unique(site_name)),
littlerange = range(logYield)[2]-range(logYield)[1],
littlesd = sd(logYield),
littlevar = var(logYield)) %>%
ungroup() %>%
left_join(varbig) %>%
mutate(pe_range = ((littlerange-bigrange)/bigrange),
pe_sd = ((littlesd-bigsd)/bigsd),
pe_var = ((littlevar-bigvar)/bigvar)) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield, totalyield_z, totalyield_sum, totalyield_sum_z, tyz_perc, tyz_sum_perc)) %>%
filter(WaterYear %in% wtryrs) %>%
mutate(wylab = substr(WaterYear, 3, 4)) %>%
arrange(totalyield_sum_z)
# calculate annual median summer flow
annmed <- tempdat %>%
#filter(WaterYear %in% c(lowyr, highyr)) %>%
group_by(site_name, WaterYear) %>%
summarize(medlogYield = median(logYield, na.rm = TRUE)) %>%
ungroup()
# arrange years by total annual yield
wtryrs_arranged <- as.numeric(unlist(pedat %>% select(WaterYear)))
# plot it
plotlist_ye <- list()
for(i in 1:length(wtryrs_arranged)) {
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i]))
mylittleg_sheds <- sheds %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i])) %>% arrange(desc(area_sqmi))
plotlist_ye[[i]] <- local({
i <- i
ggplot() +
geom_sf(data = st_as_sf(mysheds), color = "black", fill = "white", linewidth = 0.4) +
geom_sf(data = mylittleg_sheds, aes(fill = medlogYield), color = "black") +
scale_fill_viridis(option = "G", direction = 1, limits = range(annmed$medlogYield), na.value = "grey") +
geom_sf(data = st_as_sf(mynet), color = "white", linewidth = 1, lineend = "round") +
geom_sf(data = st_as_sf(mynet), color = "royalblue4", linewidth = 0.6, lineend = "round") +
geom_sf(data = mylittleg, shape = 21, fill = "white", size = 2) +
labs(fill = "Median\nsummer\nlog(Yield)") + #annotation_scale() +
coord_sf(xlim = range(st_coordinates(mylittleg_sheds)[,1]), ylim = range(st_coordinates(mylittleg_sheds)[,2])) +
theme_bw() + theme(axis.title.x = element_blank(), axis.title.y = element_blank(), legend.position = "none", axis.text = element_blank()) +
annotate("label", x = -Inf, y = -Inf, label = paste(pedat$WaterYear[i], " (", pedat$tyz_sum_perc[i], "%)", sep = ""), vjust = -0.15, hjust = -0.05, size = 5)})
}
ggarrange(plotlist = plotlist_ye, common.legend = TRUE, legend = "right", nrow = 2, ncol = 4)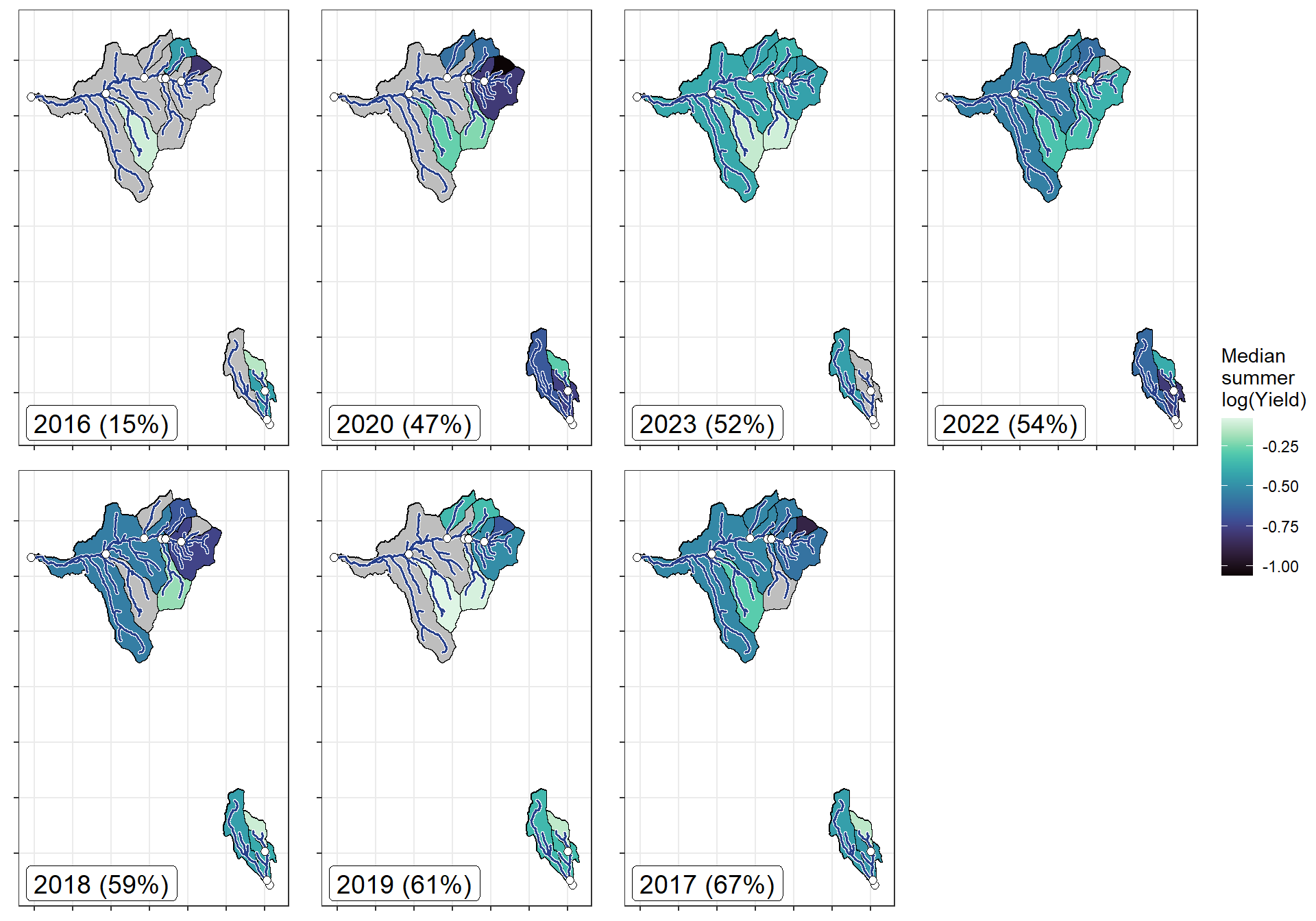
bas <- "Snake River"
orderr <- snakeorder
wtryrs <- c(2016:2024)
mysheds <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/Snake_Watersheds.shp")
mysheds <- mysheds[mysheds$site_id == "SP11",]
mynet <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams/Snake_Streams.shp")
crs(mynet) <- crs(mysheds)
mynet <- crop(mynet, mysheds)
# get lakes
lakes <- get_waterbodies(AOI = siteinfo_sp %>% filter(site_name == "Spread Creek Dam"), buffer = 100000)
lakes <- st_transform(lakes, crs(mysheds))
lakes <- st_intersection(lakes, st_as_sf(mysheds))
lakes <- lakes %>% filter(gnis_name %in% c("Leidy Lake"))
# points
mylittleg <- siteinfo_sp %>% filter(site_name %in% snakeorder) %>% mutate(site_name = factor(site_name, levels = snakeorder))
# edit geometry to reduce overlap
st_geometry(mylittleg)[mylittleg$site_name == "SF Spread Creek Lower NWIS"] <- st_point(c(-110.32226, 43.76118))
st_geometry(mylittleg)[mylittleg$site_name == "NF Spread Creek Lower"] <- st_point(c(-110.3199, 43.766533))
st_geometry(mylittleg)[mylittleg$site_name == "Grizzly Creek"] <- st_point(c(-110.23289, 43.77433))
st_geometry(mylittleg)[mylittleg$site_name == "NF Spread Creek Upper"] <- st_point(c(-110.23405, 43.77227))
st_geometry(mylittleg)[mylittleg$site_name == "SF Spread Creek Upper"] <- st_point(c(-110.31475, 43.73661))
# filter data
tempdat <- dat_clean %>%
filter(basin == bas, !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = orderr))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% unique(tempdat$WaterYear))
# calculate variability for reference gage
varbig <- tempdat_big %>%
group_by(WaterYear) %>%
summarize(bigrange = range(logYield)[2]-range(logYield)[1],
bigsd = sd(logYield),
bigvar = var(logYield)) %>%
ungroup()
# calculate total summer water availability from reference gage
#summerflow <- tempdat_big %>% group_by(WaterYear) %>% summarize(summeryield = sum(Yield_mm), summeryield_log = log(sum(Yield_mm))) %>% ungroup()
#wateravail2 <- wateravail %>% select(basin, WaterYear, totalyield, totalyield_z)
# calculate relative variation
pedat <- tempdat %>%
group_by(basin, WaterYear) %>%
summarize(nsites = length(unique(site_name)),
littlerange = range(logYield)[2]-range(logYield)[1],
littlesd = sd(logYield),
littlevar = var(logYield)) %>%
ungroup() %>%
left_join(varbig) %>%
mutate(pe_range = ((littlerange-bigrange)/bigrange),
pe_sd = ((littlesd-bigsd)/bigsd),
pe_var = ((littlevar-bigvar)/bigvar)) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield, totalyield_z, totalyield_sum, totalyield_sum_z, tyz_perc, tyz_sum_perc)) %>%
filter(WaterYear %in% wtryrs) %>%
mutate(wylab = substr(WaterYear, 3, 4)) %>%
arrange(totalyield_sum_z)
# calculate annual median summer flow
annmed <- tempdat %>%
#filter(WaterYear %in% c(lowyr, highyr)) %>%
group_by(site_name, WaterYear) %>%
summarize(medlogYield = median(logYield, na.rm = TRUE)) %>%
ungroup()
# arrange years by total annual yield
wtryrs_arranged <- as.numeric(unlist(pedat %>% select(WaterYear)))
# plot it
plotlist_sn <- list()
for(i in 1:length(wtryrs_arranged)) {
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i]))
mylittleg_sheds <- sheds %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i])) %>% arrange(desc(area_sqmi))
plotlist_sn[[i]] <- local({
i <- i
ggplot() +
geom_sf(data = st_as_sf(mysheds), color = "black", fill = "white", linewidth = 0.4) +
geom_sf(data = mylittleg_sheds, aes(fill = medlogYield), color = "black") +
scale_fill_viridis(option = "G", direction = 1, limits = range(annmed$medlogYield), na.value = "grey") +
geom_sf(data = st_as_sf(mynet), color = "white", linewidth = 1, lineend = "round") +
geom_sf(data = st_as_sf(mynet), color = "royalblue4", linewidth = 0.6, lineend = "round") +
geom_sf(data = mylittleg, shape = 21, fill = "white", size = 2) +
labs(fill = "Median\nsummer\nlog(Yield)") + #annotation_scale() +
coord_sf(xlim = range(st_coordinates(mylittleg_sheds)[,1]), ylim = range(st_coordinates(mylittleg_sheds)[,2])) +
theme_bw() + theme(axis.title.x = element_blank(), axis.title.y = element_blank(), legend.position = "none", axis.text = element_blank()) +
annotate("label", x = -Inf, y = -Inf, label = paste(pedat$WaterYear[i], " (", pedat$tyz_sum_perc[i], "%)", sep = ""), vjust = -0.15, hjust = -0.05, size = 5)})
}
ggarrange(plotlist = plotlist_sn, common.legend = TRUE, legend = "right")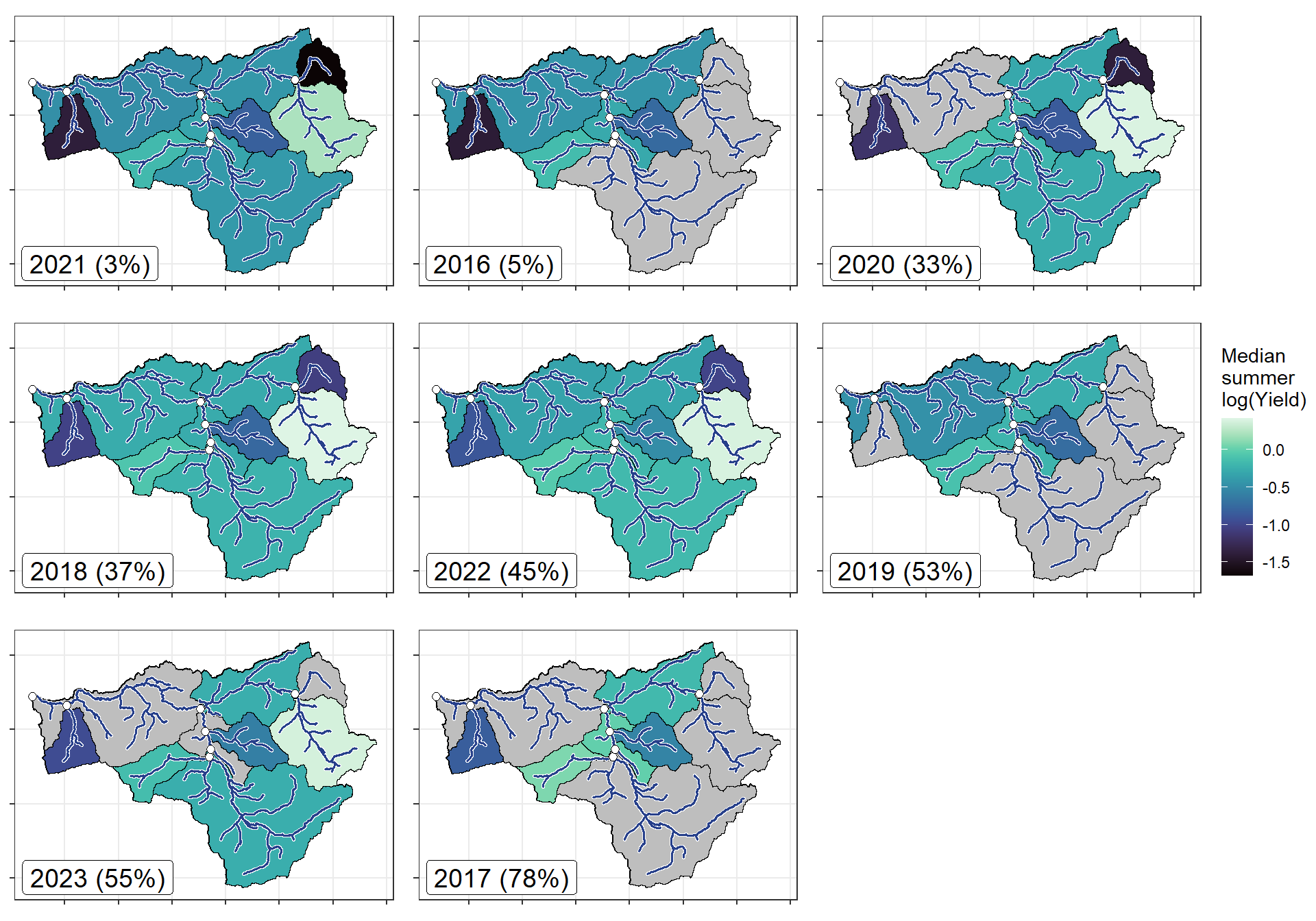
bas <- "Donner Blitzen"
orderr <- c(donnerorder, "Donner Blitzen River nr Frenchglen NWIS")
wtryrs <- c(2016:2024)
mysheds <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/Oreg_Watersheds.shp")
mysheds <- mysheds[mysheds$site_id == "DBF",]
mynet <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams/Oreg_Streams.shp")
crs(mynet) <- crs(mysheds)
mynet <- crop(mynet, mysheds)
# points
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr) %>% mutate(site_name = factor(site_name, levels = donnerorder))
# edit geometry to reduce overlap
st_geometry(mylittleg)[mylittleg$site_name == "Donner Blitzen ab Fish NWIS"] <- st_point(c(-118.84315, 42.75476))
st_geometry(mylittleg)[mylittleg$site_name == "Fish Creek"] <- st_point(c(-118.83173, 42.75911))
# filter data
tempdat <- dat_clean %>%
filter(basin == bas, !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = orderr))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% unique(tempdat$WaterYear))
# calculate variability for reference gage
varbig <- tempdat_big %>%
group_by(WaterYear) %>%
summarize(bigrange = range(logYield)[2]-range(logYield)[1],
bigsd = sd(logYield),
bigvar = var(logYield)) %>%
ungroup()
# calculate total summer water availability from reference gage
#summerflow <- tempdat_big %>% group_by(WaterYear) %>% summarize(summeryield = sum(Yield_mm), summeryield_log = log(sum(Yield_mm))) %>% ungroup()
#wateravail2 <- wateravail %>% select(basin, WaterYear, totalyield, totalyield_z)
# calculate relative variation
pedat <- tempdat %>%
group_by(basin, WaterYear) %>%
summarize(nsites = length(unique(site_name)),
littlerange = range(logYield)[2]-range(logYield)[1],
littlesd = sd(logYield),
littlevar = var(logYield)) %>%
ungroup() %>%
left_join(varbig) %>%
mutate(pe_range = ((littlerange-bigrange)/bigrange),
pe_sd = ((littlesd-bigsd)/bigsd),
pe_var = ((littlevar-bigvar)/bigvar)) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield, totalyield_z, totalyield_sum, totalyield_sum_z, tyz_perc, tyz_sum_perc)) %>%
filter(WaterYear %in% wtryrs) %>%
mutate(wylab = substr(WaterYear, 3, 4)) %>%
arrange(totalyield_sum_z)
# calculate annual median summer flow
annmed <- tempdat %>%
group_by(site_name, WaterYear) %>%
summarize(medlogYield = median(logYield, na.rm = TRUE)) %>%
ungroup() %>%
bind_rows(tempdat_big %>%
group_by(site_name, WaterYear) %>%
summarize(medlogYield = median(logYield, na.rm = TRUE)) %>%
ungroup())
# arrange years by total annual yield
wtryrs_arranged <- as.numeric(unlist(pedat %>% select(WaterYear)))
# plot it
plotlist_db <- list()
for(i in 1:length(wtryrs_arranged)) {
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i]))
mylittleg_sheds <- sheds %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i])) %>% arrange(desc(area_sqmi))
plotlist_db[[i]] <- local({
i <- i
ggplot() +
geom_sf(data = st_as_sf(mysheds), color = "black", fill = "white", linewidth = 0.4) +
geom_sf(data = mylittleg_sheds, aes(fill = medlogYield), color = "black") +
scale_fill_viridis(option = "G", direction = 1, limits = range(annmed$medlogYield), na.value = "grey") +
geom_sf(data = st_as_sf(mynet), color = "white", linewidth = 1, lineend = "round") +
geom_sf(data = st_as_sf(mynet), color = "royalblue4", linewidth = 0.6, lineend = "round") +
geom_sf(data = mylittleg, shape = 21, fill = "white", size = 2) +
labs(fill = "Median\nsummer\nlog(Yield)") + #annotation_scale() +
coord_sf(xlim = range(st_coordinates(mylittleg_sheds)[,1]), ylim = range(st_coordinates(mylittleg_sheds)[,2])) +
theme_bw() + theme(axis.title.x = element_blank(), axis.title.y = element_blank(), legend.position = "none", axis.text = element_blank()) +
annotate("label", x = -Inf, y = -Inf, label = paste(pedat$WaterYear[i], " (", pedat$tyz_sum_perc[i], "%)", sep = ""), vjust = -0.15, hjust = -0.05, size = 5)})
}
ggarrange(plotlist = plotlist_db, common.legend = TRUE, legend = "right")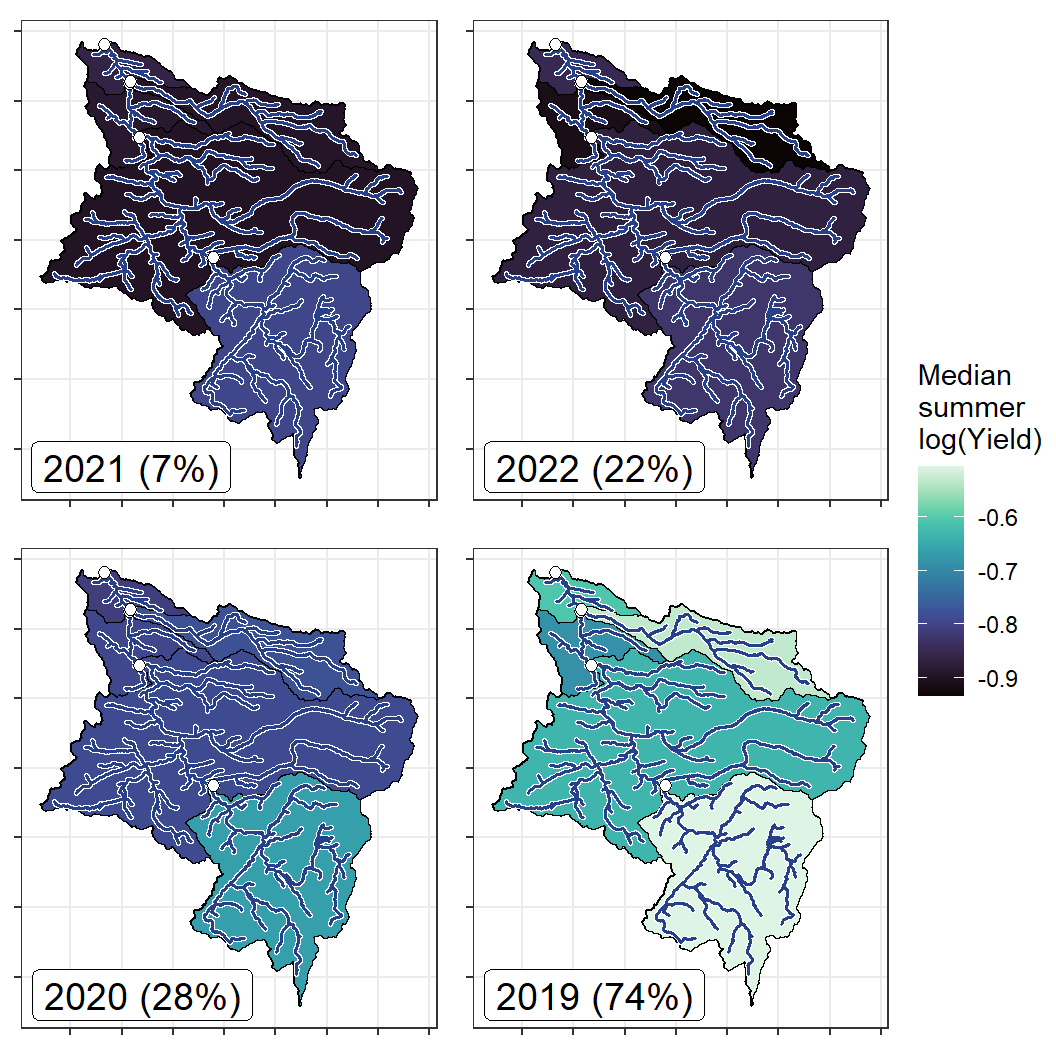
2020/2021 summer flow distributions
bas <- "West Brook"
orderr <- wborder
wtryrs <- c(2020,2021)
# filter data
tempdat <- dat_clean %>%
filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% wtryrs) %>%
mutate(site_name = factor(site_name, levels = orderr))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% unique(tempdat$WaterYear))
# annual
pann <- ggplot() +
geom_density(data = tempdat, aes(x = logYield, y = ..scaled.., color = site_name, fill = site_name), size = 0.8) +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_fill_manual(values = alpha(cet_pal(nsites, name = "i1"), 0.1)) +
geom_density(data = tempdat_big, aes(x = logYield, y = ..scaled..), color = "grey40", fill = alpha("grey40", 0.2), size = 0.8) +
xlab("Summer log(Yield, mm/day)") + ylab("Density") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.text.y = element_blank(), axis.ticks.y = element_blank(),
axis.text = element_text(color = "black"), legend.position = "none",
strip.background = element_blank(), strip.text = element_blank()) +
facet_wrap(~factor(WaterYear, levels = as.numeric(unlist(pedat %>% arrange(totalyield_sum_z) %>% select(WaterYear)))), ncol = 1)
print(pann)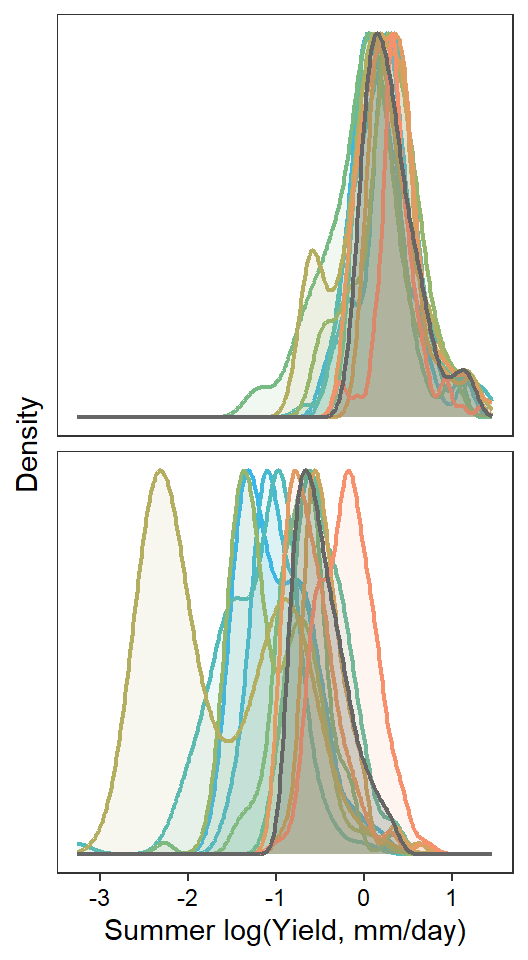
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_summerdensity_WB_annual.jpg", width = 2.75, height = 5, units = "in", res = 1000)
pann
dev.off()png
2 2020/2021 summer flow distributions
bas <- "West Brook"
orderr <- wborder
wtryrs <- c(2020,2021)
# sheds and network
mysheds <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/Mass_Watersheds.shp")
mysheds <- mysheds[mysheds$site_id == "WBR",]
mynet <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams/Mass_Streams.shp")
crs(mynet) <- crs(mysheds)
mynet <- crop(mynet, mysheds)
# get lakes
lakes <- get_waterbodies(AOI = siteinfo_sp %>% filter(site_name == "West Brook NWIS"), buffer = 10000)
lakes <- lakes %>% filter(gnis_name %in% c("Northampton Reservoir Upper", "Northampton Reservoir"))
# little g points
mylittleg <- siteinfo_sp %>% filter(site_name %in% wborder) %>% mutate(site_name = factor(site_name, levels = wborder))
# filter data
tempdat <- dat_clean %>%
filter(basin == bas, !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = orderr))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% unique(tempdat$WaterYear))
# calculate variability for reference gage
varbig <- tempdat_big %>%
group_by(WaterYear) %>%
summarize(bigrange = range(logYield)[2]-range(logYield)[1],
bigsd = sd(logYield),
bigvar = var(logYield)) %>%
ungroup()
# calculate total summer water availability from reference gage
#summerflow <- tempdat_big %>% group_by(WaterYear) %>% summarize(summeryield = sum(Yield_mm), summeryield_log = log(sum(Yield_mm))) %>% ungroup()
#wateravail2 <- wateravail %>% select(basin, WaterYear, totalyield, totalyield_z)
# calculate relative variation
pedat <- tempdat %>%
group_by(basin, WaterYear) %>%
summarize(nsites = length(unique(site_name)),
littlerange = range(logYield)[2]-range(logYield)[1],
littlesd = sd(logYield),
littlevar = var(logYield)) %>%
ungroup() %>%
left_join(varbig) %>%
mutate(pe_range = ((littlerange-bigrange)/bigrange),
pe_sd = ((littlesd-bigsd)/bigsd),
pe_var = ((littlevar-bigvar)/bigvar)) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield, totalyield_z, totalyield_sum, totalyield_sum_z, tyz_perc, tyz_sum_perc)) %>%
filter(WaterYear %in% wtryrs) %>%
mutate(wylab = substr(WaterYear, 3, 4)) %>%
arrange(totalyield_sum_z)
# calculate annual median summer flow
annmed <- tempdat %>%
#filter(WaterYear %in% c(lowyr, highyr)) %>%
group_by(site_name, WaterYear) %>%
summarize(medlogYield = median(logYield, na.rm = TRUE)) %>%
ungroup()
# arrange years by total annual yield
wtryrs_arranged <- as.numeric(unlist(pedat %>% select(WaterYear)))
# plot it
plotlist_wb <- list()
for(i in 1:length(wtryrs_arranged)) {
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i]))
mylittleg_sheds <- sheds %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i])) %>% arrange(desc(area_sqmi))
plotlist_wb[[i]] <- local({
i <- i
ggplot() +
geom_sf(data = st_as_sf(mysheds), color = "black", fill = "white", linewidth = 0.4) +
geom_sf(data = mylittleg_sheds, aes(fill = medlogYield), color = "black") +
scale_fill_viridis(option = "G", direction = 1, limits = range(annmed$medlogYield), na.value = "grey") +
geom_sf(data = st_as_sf(mynet), color = "white", linewidth = 1, lineend = "round") +
geom_sf(data = st_as_sf(mynet), color = "royalblue4", linewidth = 0.6, lineend = "round") +
geom_sf(data = lakes, color = "white", fill = "lightskyblue1", linewidth = 0.7) +
geom_sf(data = lakes, color = "royalblue4", fill = "lightskyblue1", linewidth = 0.5) +
geom_sf(data = mylittleg, shape = 21, fill = "white", size = 2) +
labs(fill = "Median\nsummer\nlog(Yield)") + #annotation_scale() +
coord_sf(xlim = range(st_coordinates(mylittleg_sheds)[,1]), ylim = range(st_coordinates(mylittleg_sheds)[,2])) +
theme_bw() + theme(axis.title.x = element_blank(), axis.title.y = element_blank(), legend.position = "none", axis.text = element_blank()) +
theme_void() + scale_x_continuous(expand = c(0.003, 0.003)) + scale_y_continuous(expand = c(0.002, 0.002))
#annotate("label", x = Inf, y = Inf, label = paste(pedat$WaterYear[i], " (", pedat$tyz_sum_perc[i], "%)", sep = ""), vjust = 1.15, hjust = 1.05, size = 5)
})
}
p <- ggarrange(plotlist = plotlist_wb, common.legend = TRUE, legend = "right", nrow = 2)
print(p)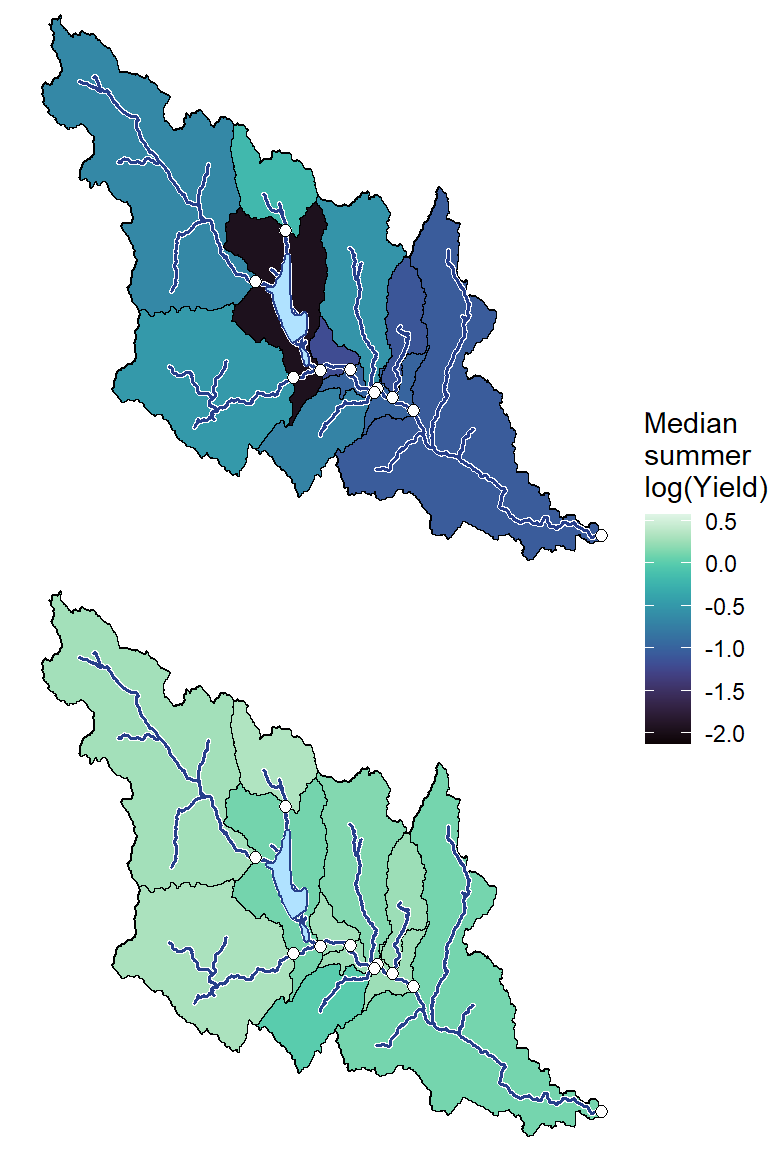
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_summermedianmap_WB_annual.jpg", width = 4, height = 6, units = "in", res = 1000)
p
dev.off()png
2 2021/2022 summer flow distributions
bas <- "Snake River"
orderr <- snakeorder
wtryrs <- c(2021,2022)
# filter data
tempdat <- dat_clean %>%
filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% wtryrs) %>%
mutate(site_name = factor(site_name, levels = orderr))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% unique(tempdat$WaterYear))
# annual
pann <- ggplot() +
geom_density(data = tempdat, aes(x = logYield, y = ..scaled.., color = site_name, fill = site_name), size = 0.8) +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
scale_fill_manual(values = alpha(cet_pal(nsites, name = "i1"), 0.1)) +
geom_density(data = tempdat_big, aes(x = logYield, y = ..scaled..), color = "grey40", fill = alpha("grey40", 0.2), size = 0.8) +
xlab("Summer log(Yield, mm/day)") + ylab("Density") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.text.y = element_blank(), axis.ticks.y = element_blank(),
axis.text = element_text(color = "black"), legend.position = "none",
strip.background = element_blank(), strip.text = element_blank()) +
facet_wrap(~factor(WaterYear, levels = as.numeric(unlist(pedat %>% arrange(totalyield_sum_z) %>% select(WaterYear)))), ncol = 1)
print(pann)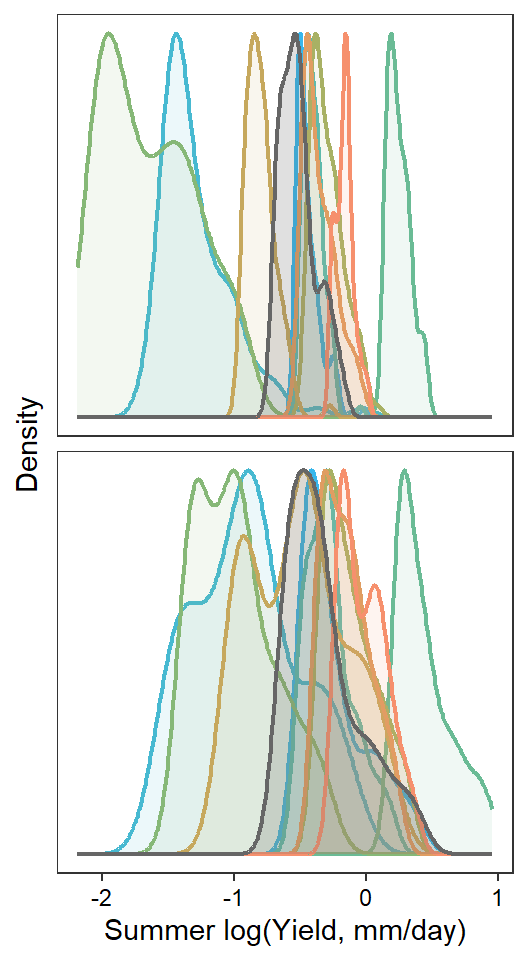
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_summerdensity_Snake_annual.jpg", width = 2.75, height = 5, units = "in", res = 1000)
pann
dev.off()png
2 2022/2022 summer flow distributions
bas <- "Snake River"
orderr <- snakeorder
wtryrs <- c(2021,2022)
mysheds <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/Snake_Watersheds.shp")
mysheds <- mysheds[mysheds$site_id == "SP11",]
mynet <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams/Snake_Streams.shp")
crs(mynet) <- crs(mysheds)
mynet <- crop(mynet, mysheds)
# get lakes
lakes <- get_waterbodies(AOI = siteinfo_sp %>% filter(site_name == "Spread Creek Dam"), buffer = 100000)
lakes <- st_transform(lakes, crs(mysheds))
lakes <- st_intersection(lakes, st_as_sf(mysheds))
lakes <- lakes %>% filter(gnis_name %in% c("Leidy Lake"))
# points
mylittleg <- siteinfo_sp %>% filter(site_name %in% snakeorder) %>% mutate(site_name = factor(site_name, levels = snakeorder))
# edit geometry to reduce overlap
st_geometry(mylittleg)[mylittleg$site_name == "SF Spread Creek Lower NWIS"] <- st_point(c(-110.32226, 43.76118))
st_geometry(mylittleg)[mylittleg$site_name == "NF Spread Creek Lower"] <- st_point(c(-110.3199, 43.766533))
st_geometry(mylittleg)[mylittleg$site_name == "Grizzly Creek"] <- st_point(c(-110.23289, 43.77433))
st_geometry(mylittleg)[mylittleg$site_name == "NF Spread Creek Upper"] <- st_point(c(-110.23405, 43.77227))
st_geometry(mylittleg)[mylittleg$site_name == "SF Spread Creek Upper"] <- st_point(c(-110.31475, 43.73661))
# filter data
tempdat <- dat_clean %>%
filter(basin == bas, !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = orderr))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% unique(tempdat$WaterYear))
# calculate variability for reference gage
varbig <- tempdat_big %>%
group_by(WaterYear) %>%
summarize(bigrange = range(logYield)[2]-range(logYield)[1],
bigsd = sd(logYield),
bigvar = var(logYield)) %>%
ungroup()
# calculate total summer water availability from reference gage
#summerflow <- tempdat_big %>% group_by(WaterYear) %>% summarize(summeryield = sum(Yield_mm), summeryield_log = log(sum(Yield_mm))) %>% ungroup()
#wateravail2 <- wateravail %>% select(basin, WaterYear, totalyield, totalyield_z)
# calculate relative variation
pedat <- tempdat %>%
group_by(basin, WaterYear) %>%
summarize(nsites = length(unique(site_name)),
littlerange = range(logYield)[2]-range(logYield)[1],
littlesd = sd(logYield),
littlevar = var(logYield)) %>%
ungroup() %>%
left_join(varbig) %>%
mutate(pe_range = ((littlerange-bigrange)/bigrange),
pe_sd = ((littlesd-bigsd)/bigsd),
pe_var = ((littlevar-bigvar)/bigvar)) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield, totalyield_z, totalyield_sum, totalyield_sum_z, tyz_perc, tyz_sum_perc)) %>%
filter(WaterYear %in% wtryrs) %>%
mutate(wylab = substr(WaterYear, 3, 4)) %>%
arrange(totalyield_sum_z)
# calculate annual median summer flow
annmed <- tempdat %>%
#filter(WaterYear %in% c(lowyr, highyr)) %>%
group_by(site_name, WaterYear) %>%
summarize(medlogYield = median(logYield, na.rm = TRUE)) %>%
ungroup()
# arrange years by total annual yield
wtryrs_arranged <- as.numeric(unlist(pedat %>% select(WaterYear)))
# plot it
plotlist_wb <- list()
for(i in 1:length(wtryrs_arranged)) {
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i]))
mylittleg_sheds <- sheds %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i])) %>% arrange(desc(area_sqmi))
plotlist_wb[[i]] <- local({
i <- i
ggplot() +
geom_sf(data = st_as_sf(mysheds), color = "black", fill = "white", linewidth = 0.4) +
geom_sf(data = mylittleg_sheds, aes(fill = medlogYield), color = "black") +
scale_fill_viridis(option = "G", direction = 1, limits = range(annmed$medlogYield), na.value = "grey") +
geom_sf(data = st_as_sf(mynet), color = "white", linewidth = 1, lineend = "round") +
geom_sf(data = st_as_sf(mynet), color = "royalblue4", linewidth = 0.6, lineend = "round") +
geom_sf(data = lakes, color = "white", fill = "lightskyblue1", linewidth = 0.7) +
geom_sf(data = lakes, color = "royalblue4", fill = "lightskyblue1", linewidth = 0.5) +
geom_sf(data = mylittleg, shape = 21, fill = "white", size = 2) +
labs(fill = "Median\nsummer\nlog(Yield)") + #annotation_scale() +
coord_sf(xlim = range(st_coordinates(mylittleg_sheds)[,1]), ylim = range(st_coordinates(mylittleg_sheds)[,2])) +
theme_bw() + theme(axis.title.x = element_blank(), axis.title.y = element_blank(), legend.position = "none", axis.text = element_blank()) +
theme_void() + scale_x_continuous(expand = c(0.003, 0.003)) + scale_y_continuous(expand = c(0.002, 0.002))
#annotate("label", x = Inf, y = Inf, label = paste(pedat$WaterYear[i], " (", pedat$tyz_sum_perc[i], "%)", sep = ""), vjust = 1.15, hjust = 1.05, size = 5)
})
}
p <- ggarrange(plotlist = plotlist_wb, common.legend = TRUE, legend = "right", nrow = 2)
print(p)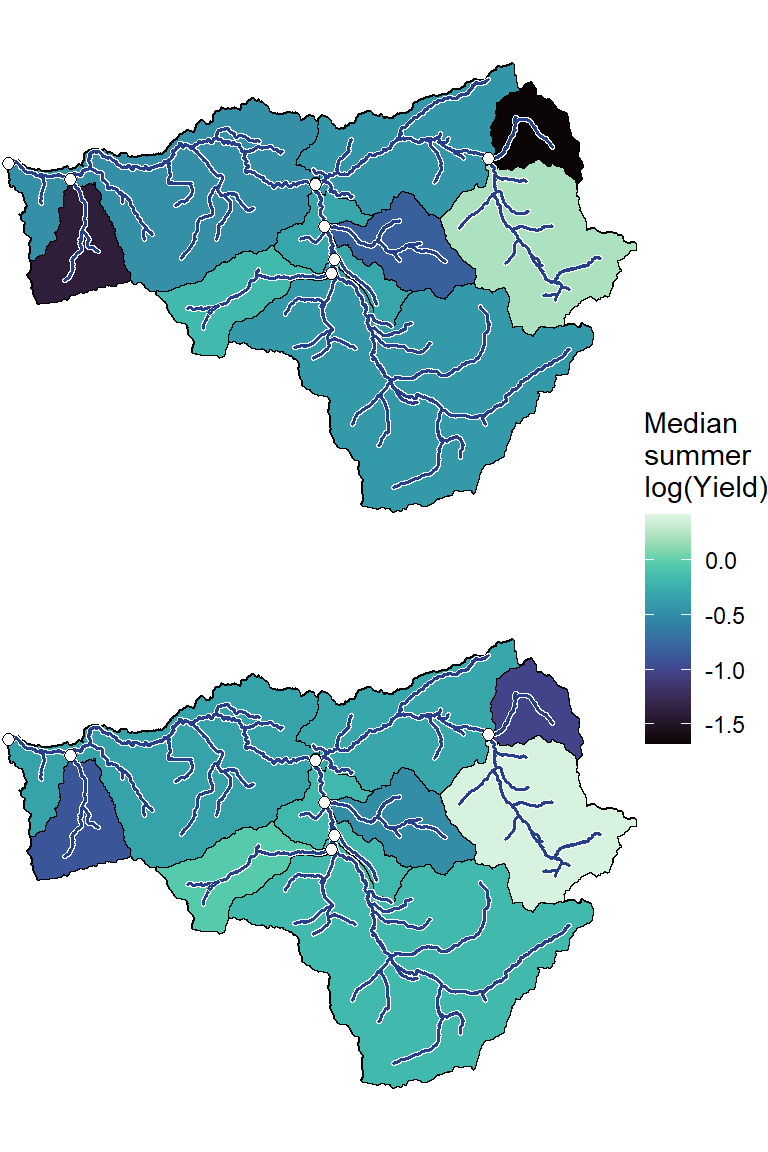
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_summermedianmap_Snake_annual.jpg", width = 4, height = 6, units = "in", res = 1000)
p
dev.off()png
2 Combine interannual distributions of summer flow, scatterplots showing relationship between water availability and relative range among little g’s, and maps of median summer flow in a low and high water year
Redraw Yellowstone to move upper Shields and Duck Creek closer together
bas <- "Shields River"
orderr <- yellorder
wtryrs <- c(2016:2020,2022:2024)
mysheds <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/Shields_Watersheds.shp")
mysheds <- mysheds[mysheds$site_id %in% c("SRS", "DU01"),]
mynet <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams/Shields_Streams.shp")
crs(mynet) <- crs(mysheds)
mynet <- crop(mynet, mysheds)
# points
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr) %>% mutate(site_name = factor(site_name, levels = orderr)) #%>% filter(subbasin != "Duck Creek")
# filter data
tempdat <- dat_clean %>%
filter(basin == bas, !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = orderr))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% unique(tempdat$WaterYear))
# calculate variability for reference gage
varbig <- tempdat_big %>%
group_by(WaterYear) %>%
summarize(bigrange = range(logYield)[2]-range(logYield)[1],
bigsd = sd(logYield),
bigvar = var(logYield)) %>%
ungroup()
# calculate total summer water availability from reference gage
#summerflow <- tempdat_big %>% group_by(WaterYear) %>% summarize(summeryield = sum(Yield_mm), summeryield_log = log(sum(Yield_mm))) %>% ungroup()
#wateravail2 <- wateravail %>% select(basin, WaterYear, totalyield, totalyield_z)
# calculate relative variation
pedat <- tempdat %>%
group_by(basin, WaterYear) %>%
summarize(nsites = length(unique(site_name)),
littlerange = range(logYield)[2]-range(logYield)[1],
littlesd = sd(logYield),
littlevar = var(logYield)) %>%
ungroup() %>%
left_join(varbig) %>%
mutate(pe_range = ((littlerange-bigrange)/bigrange),
pe_sd = ((littlesd-bigsd)/bigsd),
pe_var = ((littlevar-bigvar)/bigvar)) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield, totalyield_z, totalyield_sum, totalyield_sum_z, tyz_perc, tyz_sum_perc)) %>%
filter(WaterYear %in% wtryrs) %>%
mutate(wylab = substr(WaterYear, 3, 4)) %>%
arrange(totalyield_sum_z)
# calculate annual median summer flow
annmed <- tempdat %>%
#filter(WaterYear %in% c(lowyr, highyr)) %>%
group_by(site_name, WaterYear) %>%
summarize(medlogYield = median(logYield, na.rm = TRUE)) %>%
ungroup()
# arrange years by total annual yield
wtryrs_arranged <- as.numeric(unlist(pedat %>% select(WaterYear)))
# 2020
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == 2020))
mylittleg_sheds <- sheds %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == 2020)) %>% arrange(desc(area_sqmi))
mainmap <- ggplot() +
geom_sf(data = st_as_sf(mysheds), color = "black", fill = "white", linewidth = 0.4) +
geom_sf(data = mylittleg_sheds, aes(fill = medlogYield), color = "black") +
scale_fill_viridis(option = "G", direction = 1, limits = range(annmed$medlogYield), na.value = "grey") +
geom_sf(data = st_as_sf(mynet), color = "white", linewidth = 1, lineend = "round") +
geom_sf(data = st_as_sf(mynet), color = "royalblue4", linewidth = 0.6, lineend = "round") +
geom_sf(data = mylittleg, shape = 21, fill = "white", size = 2) +
labs(fill = "Median\nsummer\nlog(Yield)") + #annotation_scale() +
coord_sf(xlim = c(-110.555,-110.30), ylim = c(46.07,46.23)) +
theme_bw() + theme_void() +
theme(axis.title.x = element_blank(), axis.title.y = element_blank(), axis.text = element_blank()) +
scale_x_continuous(expand = c(0.002, 0.002)) + scale_y_continuous(expand = c(0.002, 0.002)) +
annotate("label", x = -Inf, y = Inf, label = "2020 (47%)", vjust = 1.15, hjust = -0.05, size = 5)
dbp1 <- local({
i <- i
ggdraw(mainmap + theme(legend.position = "none")) +
draw_plot(
{
ggplot() +
geom_sf(data = st_as_sf(mysheds), color = "black", fill = "white", linewidth = 0.4) +
geom_sf(data = mylittleg_sheds, aes(fill = medlogYield), color = "black") +
scale_fill_viridis(option = "G", direction = 1, limits = range(annmed$medlogYield), na.value = "grey") +
geom_sf(data = st_as_sf(mynet), color = "white", linewidth = 1, lineend = "round") +
geom_sf(data = st_as_sf(mynet), color = "royalblue4", linewidth = 0.6, lineend = "round") +
geom_sf(data = mylittleg, shape = 21, fill = "white", size = 2) +
labs(fill = "Median\nsummer\nlog(Yield)") + #annotation_scale() +
coord_sf(xlim = c(-110.31,-110.23), ylim = c(45.865,45.96)) +
theme_bw() + theme_void() + theme(legend.position = "none") +
scale_x_continuous(expand = c(0.002, 0.002)) + scale_y_continuous(expand = c(0.002, 0.002))
},
x = 0.05, y = 0, width = 0.3, height = 0.5)
})
# 2017
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == 2017))
mylittleg_sheds <- sheds %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == 2017)) %>% arrange(desc(area_sqmi))
mainmap <- ggplot() +
geom_sf(data = st_as_sf(mysheds), color = "black", fill = "white", linewidth = 0.4) +
geom_sf(data = mylittleg_sheds, aes(fill = medlogYield), color = "black") +
scale_fill_viridis(option = "G", direction = 1, limits = range(annmed$medlogYield), na.value = "grey") +
geom_sf(data = st_as_sf(mynet), color = "white", linewidth = 1, lineend = "round") +
geom_sf(data = st_as_sf(mynet), color = "royalblue4", linewidth = 0.6, lineend = "round") +
geom_sf(data = mylittleg, shape = 21, fill = "white", size = 2) +
labs(fill = "Median\nsummer\nlog(Yield)") + #annotation_scale() +
coord_sf(xlim = c(-110.555,-110.30), ylim = c(46.07,46.23)) +
theme_bw() + theme_void() +
theme(axis.title.x = element_blank(), axis.title.y = element_blank(), axis.text = element_blank()) +
scale_x_continuous(expand = c(0.002, 0.002)) + scale_y_continuous(expand = c(0.002, 0.002)) +
annotate("label", x = -Inf, y = Inf, label = "2017 (67%)", vjust = 1.15, hjust = -0.05, size = 5)
dbp2 <- local({
i <- i
ggdraw(mainmap + theme(legend.position = "none")) +
draw_plot(
{
ggplot() +
geom_sf(data = st_as_sf(mysheds), color = "black", fill = "white", linewidth = 0.4) +
geom_sf(data = mylittleg_sheds, aes(fill = medlogYield), color = "black") +
scale_fill_viridis(option = "G", direction = 1, limits = range(annmed$medlogYield), na.value = "grey") +
geom_sf(data = st_as_sf(mynet), color = "white", linewidth = 1, lineend = "round") +
geom_sf(data = st_as_sf(mynet), color = "royalblue4", linewidth = 0.6, lineend = "round") +
geom_sf(data = mylittleg, shape = 21, fill = "white", size = 2) +
labs(fill = "Median\nsummer\nlog(Yield)") + #annotation_scale() +
coord_sf(xlim = c(-110.31,-110.23), ylim = c(45.865,45.96)) +
theme_bw() + theme_void() + theme(legend.position = "none") +
scale_x_continuous(expand = c(0.002, 0.002)) + scale_y_continuous(expand = c(0.002, 0.002))
},
x = 0.05, y = 0, width = 0.3, height = 0.5)
})
#ggarrange(dbp1, dbp2, as_ggplot(ggpubr::get_legend(mainmap)), nrow = 1, widths = c(1,1,0.2))Get pieces
# West Brook
p1 <- ggarrange(portfolioplot(bas = "West Brook", orderr = wborder, type = "interannual", wtryrs = c(2020:2024)) + theme(axis.title.x = element_blank()),
portfolioplot(bas = "West Brook", orderr = wborder, type = "scatter", wtryrs = c(2020:2024)) + theme(axis.title.x = element_blank()),
ggarrange(plotlist_wb[[2]] + theme_void() + scale_x_continuous(expand = c(0.002, 0.002)) + scale_y_continuous(expand = c(0.002, 0.002)) + theme(legend.position = "none"),
plotlist_wb[[4]] + theme_void() + scale_x_continuous(expand = c(0.002, 0.002)) + scale_y_continuous(expand = c(0.002, 0.002)) + theme(legend.position = "none")),
widths = c(1,1,2), align = "h", nrow = 1)
# Paine
p2 <- ggarrange(portfolioplot(bas = "Paine Run", orderr = paineorder, type = "interannual", wtryrs = c(2019:2022)) + theme(axis.title.x = element_blank()),
portfolioplot(bas = "Paine Run", orderr = paineorder, type = "scatter", wtryrs = c(2019:2022)) + theme(axis.title.x = element_blank()),
ggarrange(plotlist_pa[[1]] + theme_void() + scale_x_continuous(expand = c(0.002, 0.002)) + scale_y_continuous(expand = c(0.002, 0.002)) + theme(legend.position = "none"),
plotlist_pa[[4]] + theme_void() + scale_x_continuous(expand = c(0.002, 0.002)) + scale_y_continuous(expand = c(0.002, 0.002)) + theme(legend.position = "none")),
widths = c(1,1,2), align = "h", nrow = 1)
# Staunton
p3 <- ggarrange(portfolioplot(bas = "Staunton River", orderr = stauntorder, type = "interannual", wtryrs = c(2019:2022)) + theme(axis.title.x = element_blank()),
portfolioplot(bas = "Staunton River", orderr = stauntorder, type = "scatter", wtryrs = c(2019:2022)) + theme(axis.title.x = element_blank()),
ggarrange(plotlist_st[[1]] + theme_void() + scale_x_continuous(expand = c(0.002, 0.002)) + scale_y_continuous(expand = c(0.002, 0.002)) + theme(legend.position = "none"),
plotlist_st[[4]] + theme_void() + scale_x_continuous(expand = c(0.002, 0.002)) + scale_y_continuous(expand = c(0.002, 0.002)) + theme(legend.position = "none")),
widths = c(1,1,2), align = "h", nrow = 1)
# Flathead
p4 <- ggarrange(portfolioplot(bas = "Flathead", orderr = flatorder, type = "interannual", wtryrs = c(2018:2024)) + theme(axis.title.x = element_blank()),
portfolioplot(bas = "Flathead", orderr = flatorder, type = "scatter", wtryrs = c(2018:2024)) + theme(axis.title.x = element_blank()),
ggarrange(plotlist_fl[[2]] + theme_void() + scale_x_continuous(expand = c(0.003, 0.003)) + scale_y_continuous(expand = c(0.002, 0.002)) + theme(legend.position = "none"),
plotlist_fl[[6]] + theme_void() + scale_x_continuous(expand = c(0.003, 0.003)) + scale_y_continuous(expand = c(0.002, 0.002)) + theme(legend.position = "none")),
widths = c(1,1,2), align = "h", nrow = 1)
# Yellowstone
p5 <- ggarrange(portfolioplot(bas = "Shields River", orderr = yellorder, type = "interannual", wtryrs = c(2016:2020,2022:2024)) + theme(axis.title.x = element_blank()),
portfolioplot(bas = "Shields River", orderr = yellorder, type = "scatter", wtryrs = c(2016:2020,2022:2024)) + theme(axis.title.x = element_blank()),
ggarrange(dbp1, dbp2, nrow = 1),
widths = c(1,1,2), align = "h", nrow = 1)
# Snake River
p6 <- ggarrange(portfolioplot(bas = "Snake River", orderr = snakeorder, type = "interannual", wtryrs = c(2016:2024)) + theme(axis.title.x = element_blank()),
portfolioplot(bas = "Snake River", orderr = snakeorder, type = "scatter", wtryrs = c(2016:2024)) + theme(axis.title.x = element_blank()),
ggarrange(plotlist_sn[[1]] + theme_void() + scale_x_continuous(expand = c(0.003, 0.003)) + scale_y_continuous(expand = c(0.002, 0.002)) + theme(legend.position = "none"),
plotlist_sn[[5]] + theme_void() + scale_x_continuous(expand = c(0.003, 0.003)) + scale_y_continuous(expand = c(0.002, 0.002)) + theme(legend.position = "none")),
widths = c(1,1,2), align = "h", nrow = 1)
# Donner Blitzen
p7 <- ggarrange(portfolioplot(bas = "Donner Blitzen", orderr = donnerorder, type = "interannual", wtryrs = c(2016:2024)),
portfolioplot(bas = "Donner Blitzen", orderr = donnerorder, type = "scatter", wtryrs = c(2016:2024)),
ggarrange(plotlist_db[[1]] + theme_void() + scale_x_continuous(expand = c(0.002, 0.002)) + scale_y_continuous(expand = c(0.003, 0.003)) + theme(legend.position = "none"),
plotlist_db[[4]] + theme_void() + scale_x_continuous(expand = c(0.002, 0.002)) + scale_y_continuous(expand = c(0.003, 0.003)) + theme(legend.position = "none")),
widths = c(1,1,2), align = "h", nrow = 1)
egg::ggarrange(p1, p2, p3, p4, p5, p6, p7, nrow = 7)jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/EcoD_Obj1_bigplot.jpg", width = 12, height = 20, units = "in", res = 1000)
egg::ggarrange(p1, p2, p3, p4, p5, p6, p7, nrow = 7)
dev.off()Time series plots, West Brook and Flathead
### FLATHEAD
tempdat <- dat_clean %>%
filter(basin == "Flathead", date >= date("2018-07-29"), date <= date("2023-08-11")) %>%
mutate(site_name = factor(site_name, levels = flatorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Flathead", date >= date("2018-07-29"), date <= date("2023-08-11"))
# color
pfl1 <- ggplot() +
geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = flatorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
ylim(-3,2) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Flathead River", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
p <- annotate_figure(egg::ggarrange(pwb1, pfl1 + ylim(-2.4,1.72), ncol = 1), left = "log(Yield, mm/day)")
print(p)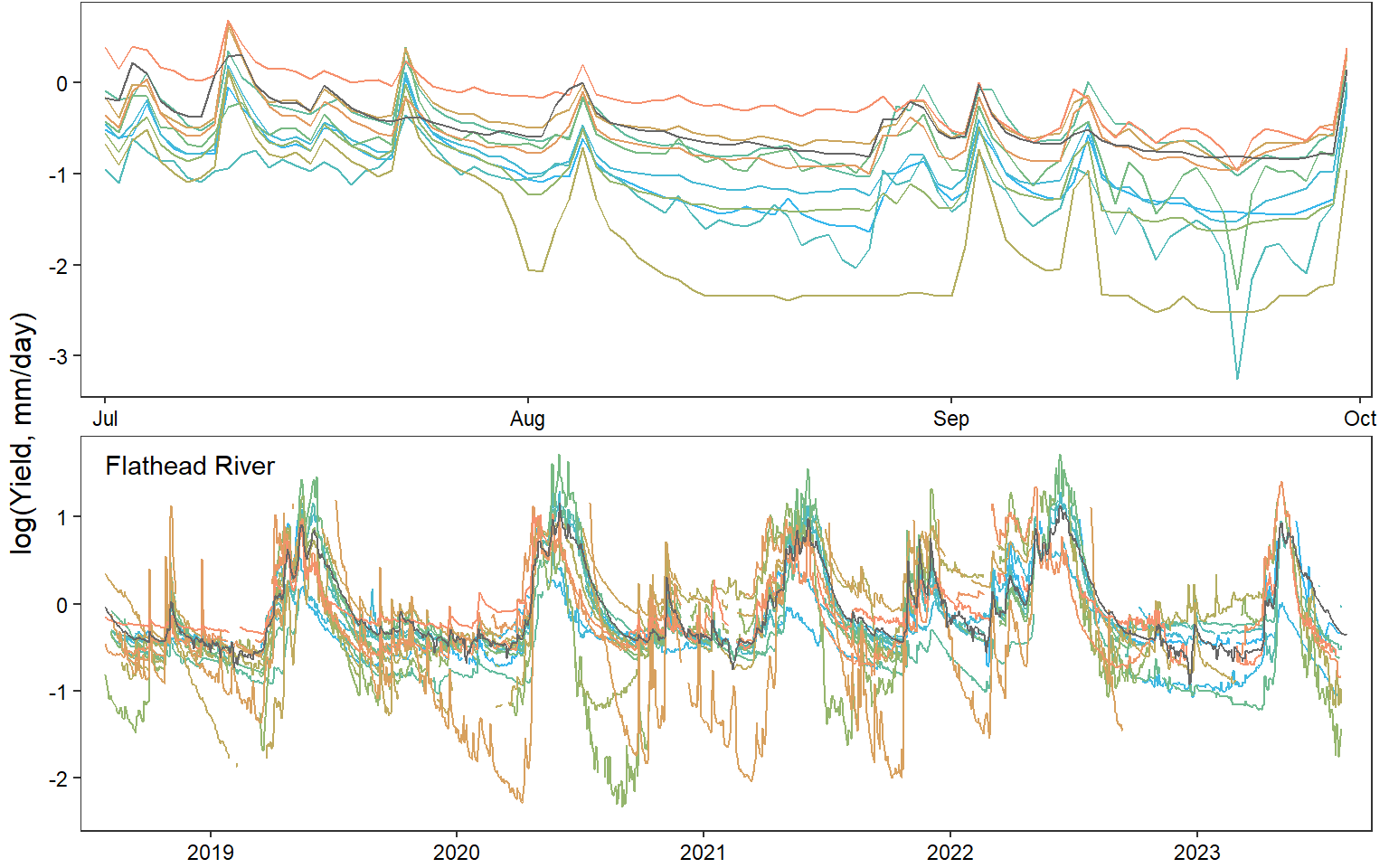
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_timeseries_WBFlat.jpg", width = 8, height = 5, units = "in", res = 1000)
p
dev.off()png
2 #### NO LITTLE Gs
### WEST BROOK
tempdat <- dat_clean %>%
filter(basin == "West Brook") %>%
mutate(site_name = factor(site_name, levels = wborder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "West Brook", date >= date("2020-01-01"), date <= date("2025-01-03"))
# color
pwb1_nlg <- ggplot() +
#geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = wborder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
ylim(-3,2) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "The West Brook", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### FLATHEAD
tempdat <- dat_clean %>%
filter(basin == "Flathead", date >= date("2018-07-29"), date <= date("2023-08-11")) %>%
mutate(site_name = factor(site_name, levels = flatorder))
tempdat <- fill_missing_dates(tempdat, dates = date, groups = site_name, pad_ends = FALSE)
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Flathead", date >= date("2018-07-29"), date <= date("2023-08-11"))
# color
pfl1_nlg <- ggplot() +
#geom_line(data = tempdat, aes(x = date, y = logYield, color = factor(site_name, levels = flatorder))) +
geom_line(data = tempdat_big, aes(x = date, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
ylim(-2.4,1.72) +
annotate("text", x = min(c(tempdat_big$date, tempdat$date), na.rm = TRUE), y = max(c(tempdat_big$logYield, tempdat$logYield), na.rm = TRUE), label = "Flathead River", hjust = 0, vjust = 1) +
scale_x_date(expand = c(0.02,0.02)) + #scale_y_continuous(expand = c(0,0)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
p <- annotate_figure(egg::ggarrange(pwb1_nlg, pfl1_nlg, ncol = 1), left = "log(Yield, mm/day)")
print(p)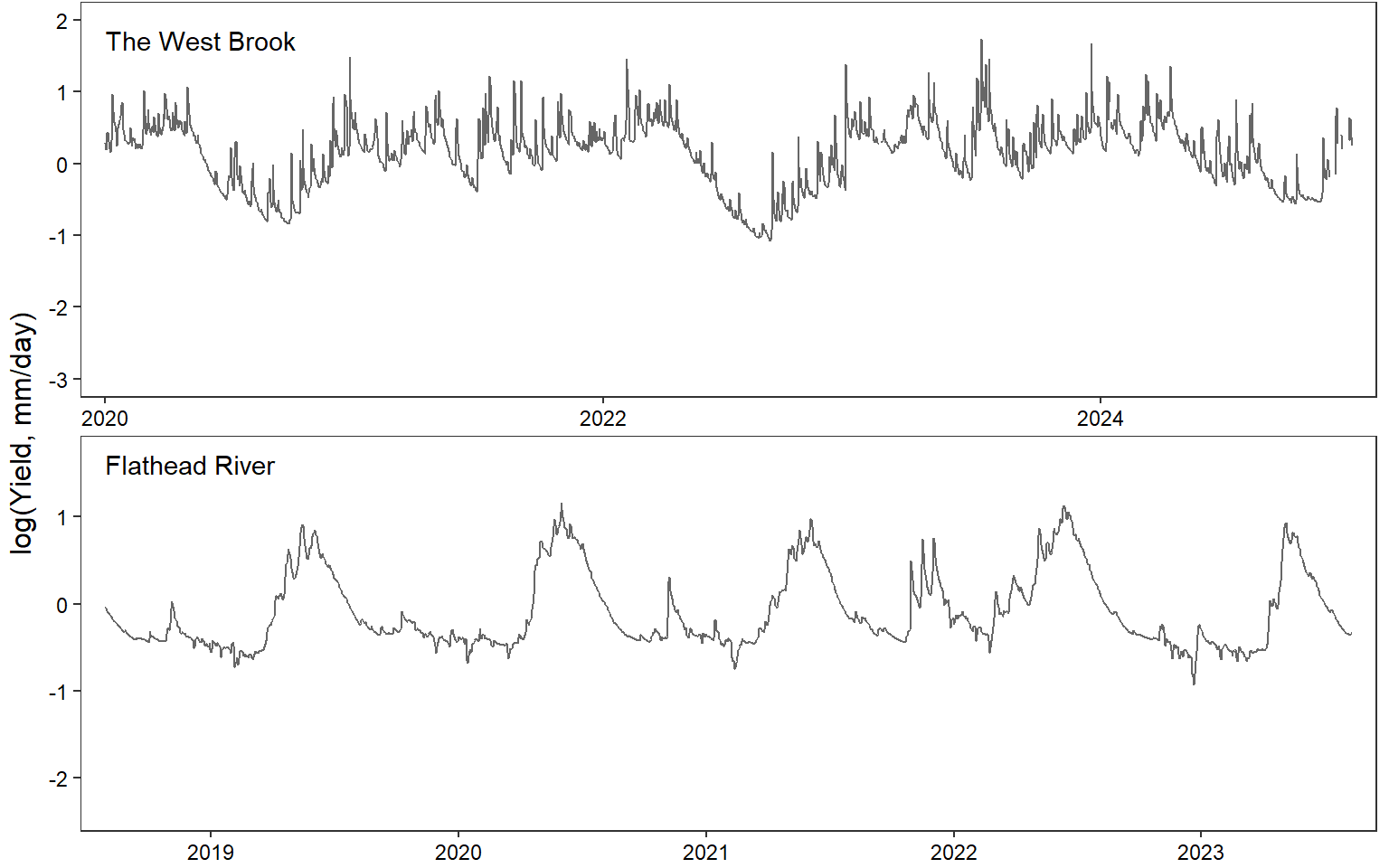
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_timeseries_WBFlat_nolittle.jpg", width = 8, height = 5, units = "in", res = 1000)
p
dev.off()png
2 Summer log(yield) distributions for West Brook and Flathead
p <- egg::ggarrange(portfolioplot(bas = "West Brook", orderr = wborder, type = "interannual", wtryrs = c(2020:2024)) + theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(), plot.margin = margin(t = 1, r = 1, b = 2, l = 1)),
portfolioplot(bas = "Flathead", orderr = flatorder, type = "interannual", wtryrs = c(2018:2024)) + theme(axis.title = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank(), plot.margin = margin(t = 1, r = 1, b = 2, l = 1)),
nrow = 2)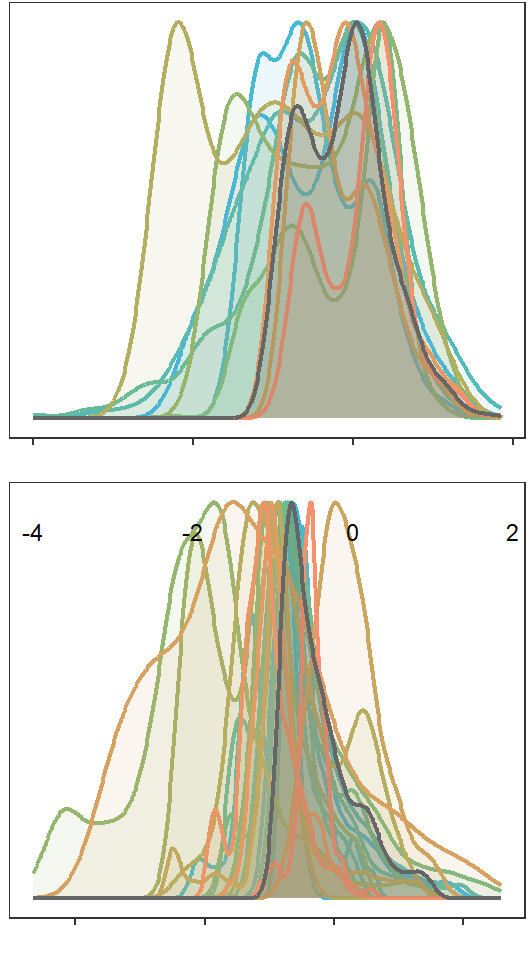
print(p)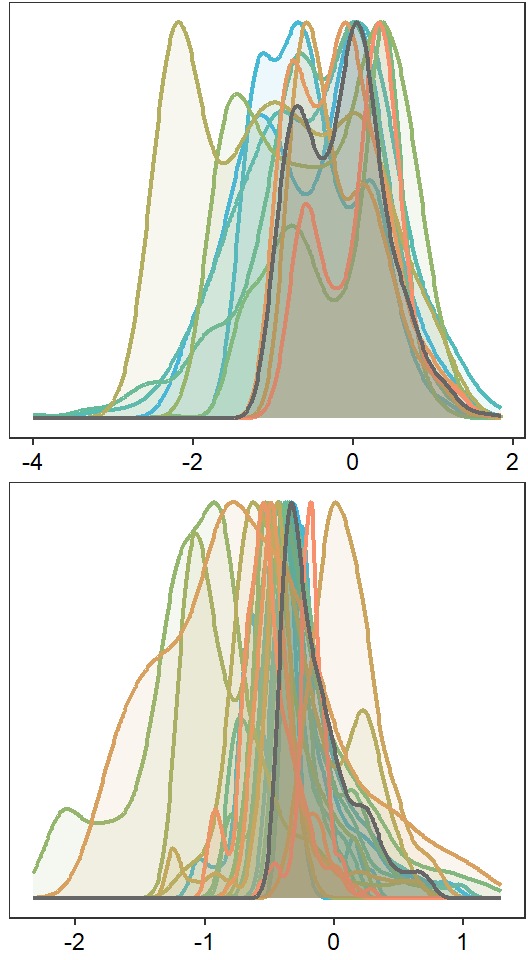
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_summerdensity_WBFlat.jpg", width = 2.75, height = 5, units = "in", res = 1000)
p
dev.off()png
2 bas <- "Snake River"
orderr <- snakeorder
wtryrs <- c(2022)
mysheds <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Watersheds/Snake_Watersheds.shp")
mysheds <- mysheds[mysheds$site_id == "SP11",]
mynet <- vect("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Watershed Delineation/Streams/Snake_Streams.shp")
crs(mynet) <- crs(mysheds)
mynet <- crop(mynet, mysheds)
# get lakes
lakes <- get_waterbodies(AOI = siteinfo_sp %>% filter(site_name == "Spread Creek Dam"), buffer = 100000)
lakes <- st_transform(lakes, crs(mysheds))
lakes <- st_intersection(lakes, st_as_sf(mysheds))
lakes <- lakes %>% filter(gnis_name %in% c("Leidy Lake"))
# points
mylittleg <- siteinfo_sp %>% filter(site_name %in% snakeorder) %>% mutate(site_name = factor(site_name, levels = snakeorder))
# edit geometry to reduce overlap
st_geometry(mylittleg)[mylittleg$site_name == "SF Spread Creek Lower NWIS"] <- st_point(c(-110.32226, 43.76118))
st_geometry(mylittleg)[mylittleg$site_name == "NF Spread Creek Lower"] <- st_point(c(-110.3199, 43.766533))
st_geometry(mylittleg)[mylittleg$site_name == "Grizzly Creek"] <- st_point(c(-110.23289, 43.77433))
st_geometry(mylittleg)[mylittleg$site_name == "NF Spread Creek Upper"] <- st_point(c(-110.23405, 43.77227))
st_geometry(mylittleg)[mylittleg$site_name == "SF Spread Creek Upper"] <- st_point(c(-110.31475, 43.73661))
# filter data
tempdat <- dat_clean %>%
filter(basin == bas, !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = orderr))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == bas, !is.na(logYield), Month %in% c(7:9), WaterYear %in% unique(tempdat$WaterYear))
# calculate variability for reference gage
varbig <- tempdat_big %>%
group_by(WaterYear) %>%
summarize(bigrange = range(logYield)[2]-range(logYield)[1],
bigsd = sd(logYield),
bigvar = var(logYield)) %>%
ungroup()
# calculate total summer water availability from reference gage
#summerflow <- tempdat_big %>% group_by(WaterYear) %>% summarize(summeryield = sum(Yield_mm), summeryield_log = log(sum(Yield_mm))) %>% ungroup()
#wateravail2 <- wateravail %>% select(basin, WaterYear, totalyield, totalyield_z)
# calculate relative variation
pedat <- tempdat %>%
group_by(basin, WaterYear) %>%
summarize(nsites = length(unique(site_name)),
littlerange = range(logYield)[2]-range(logYield)[1],
littlesd = sd(logYield),
littlevar = var(logYield)) %>%
ungroup() %>%
left_join(varbig) %>%
mutate(pe_range = ((littlerange-bigrange)/bigrange),
pe_sd = ((littlesd-bigsd)/bigsd),
pe_var = ((littlevar-bigvar)/bigvar)) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield, totalyield_z, totalyield_sum, totalyield_sum_z, tyz_perc, tyz_sum_perc)) %>%
filter(WaterYear %in% wtryrs) %>%
mutate(wylab = substr(WaterYear, 3, 4)) %>%
arrange(totalyield_sum_z)
# calculate annual median summer flow
annmed <- tempdat %>%
#filter(WaterYear %in% c(lowyr, highyr)) %>%
group_by(site_name, WaterYear) %>%
summarize(medlogYield = median(logYield, na.rm = TRUE)) %>%
ungroup()
# arrange years by total annual yield
wtryrs_arranged <- as.numeric(unlist(pedat %>% select(WaterYear)))
# plot it
plotlist_sn <- list()
for(i in 1:length(wtryrs_arranged)) {
mylittleg <- siteinfo_sp %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i]))
mylittleg_sheds <- sheds %>% filter(site_name %in% orderr) %>% left_join(annmed %>% filter(WaterYear == wtryrs_arranged[i])) %>% arrange(desc(area_sqmi))
plotlist_sn[[i]] <- local({
i <- i
ggplot() +
geom_sf(data = st_as_sf(mysheds), color = "black", fill = "white", linewidth = 0.4) +
geom_sf(data = mylittleg_sheds, aes(fill = medlogYield), color = "black") +
scale_fill_viridis(option = "A", direction = 1, limits = range(annmed$medlogYield), na.value = "grey") +
geom_sf(data = st_as_sf(mynet), color = "white", linewidth = 1, lineend = "round") +
geom_sf(data = st_as_sf(mynet), color = "royalblue4", linewidth = 0.6, lineend = "round") +
geom_sf(data = mylittleg, shape = 21, fill = "white", size = 2) +
labs(fill = "Median\nsummer\nlog(Yield)") + #annotation_scale() +
coord_sf(xlim = range(st_coordinates(mylittleg_sheds)[,1]), ylim = range(st_coordinates(mylittleg_sheds)[,2])) +
theme_bw() + theme(axis.title.x = element_blank(), axis.title.y = element_blank(), legend.position = "none", axis.text = element_blank()) +
theme_void() + scale_x_continuous(expand = c(0.003, 0.003)) + scale_y_continuous(expand = c(0.002, 0.002))
# annotate("label", x = -Inf, y = -Inf, label = paste(pedat$WaterYear[i], " (", pedat$tyz_sum_perc[i], "%)", sep = ""), vjust = -0.15, hjust = -0.05, size = 5)
})
}
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Presentation figs/EcoD_summermedianmap_Snake2021.jpg", width = 4, height = 3, units = "in", res = 1000)
ggarrange(plotlist = plotlist_sn, common.legend = TRUE, legend = "right")
dev.off()png
2 For each basin, use ridgeline plots to show distribution of daily flow values and monthly means by month, year, and site.
myridgesfun <- function(bas, orderr) {
nsites <- length(orderr)
# daily data
td <- dat_clean %>% filter(basin == bas) %>%
mutate(MonthName = factor(MonthName, levels = rev(c("Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sep", "Oct", "Nov", "Dec"))),
CalendarYear = factor(CalendarYear))
# summarize by month
td2 <- td %>%
group_by(subbasin, site_name, designation, CalendarYear, Month, MonthName) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
mutate(MonthName = factor(MonthName, levels = rev(c("Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sep", "Oct", "Nov", "Dec"))),
CalendarYear = factor(CalendarYear))
# big g data
tempdat_big <- dat_clean_big %>%
filter(basin == bas, date >= min(td$date), date <= max(td$date)) %>%
group_by(site_name, CalendarYear, Month, MonthName) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
mutate(MonthName = factor(MonthName, levels = rev(c("Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sep", "Oct", "Nov", "Dec"))),
CalendarYear = factor(CalendarYear))
return(ggplot(data = td2) +
geom_density_ridges(data = td, aes(x = logYield, y = MonthName), alpha = 0.5) +
geom_line(data = td2, aes(x = logYield, y = MonthName, group = site_name, color = factor(site_name, levels = orderr)), orientation = "y", alpha = 0.4) +
geom_point(data = td2, aes(x = logYield, y = MonthName, group = site_name, color = factor(site_name, levels = orderr)), alpha = 0.4) +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
geom_line(data = tempdat_big, aes(x = logYield, y = MonthName, group = site_name), orientation = "y", alpha = 0.5) +
geom_point(data = tempdat_big, aes(x = logYield, y = MonthName, group = site_name), alpha = 0.5) +
theme_bw() + theme(legend.position = "none", panel.grid.major = element_blank(), panel.grid.minor = element_blank()) +
facet_wrap2(~CalendarYear, nrow = 3, ncol = 3, trim_blank = FALSE) +
xlab("log(Yield, mm/day)") + ylab(""))
}myridgesfun(bas = "West Brook", orderr = wborder)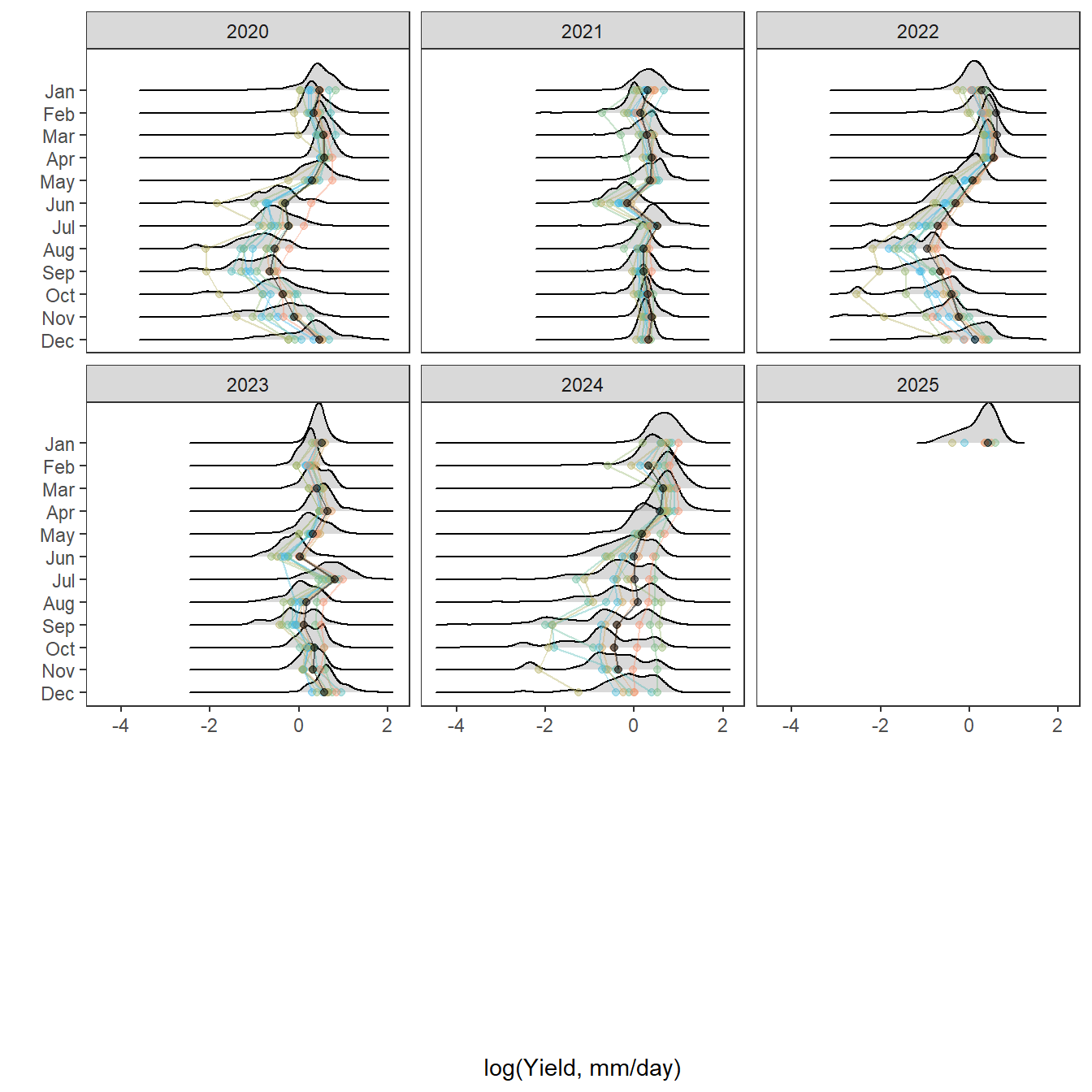
myridgesfun(bas = "Paine Run", orderr = paineorder)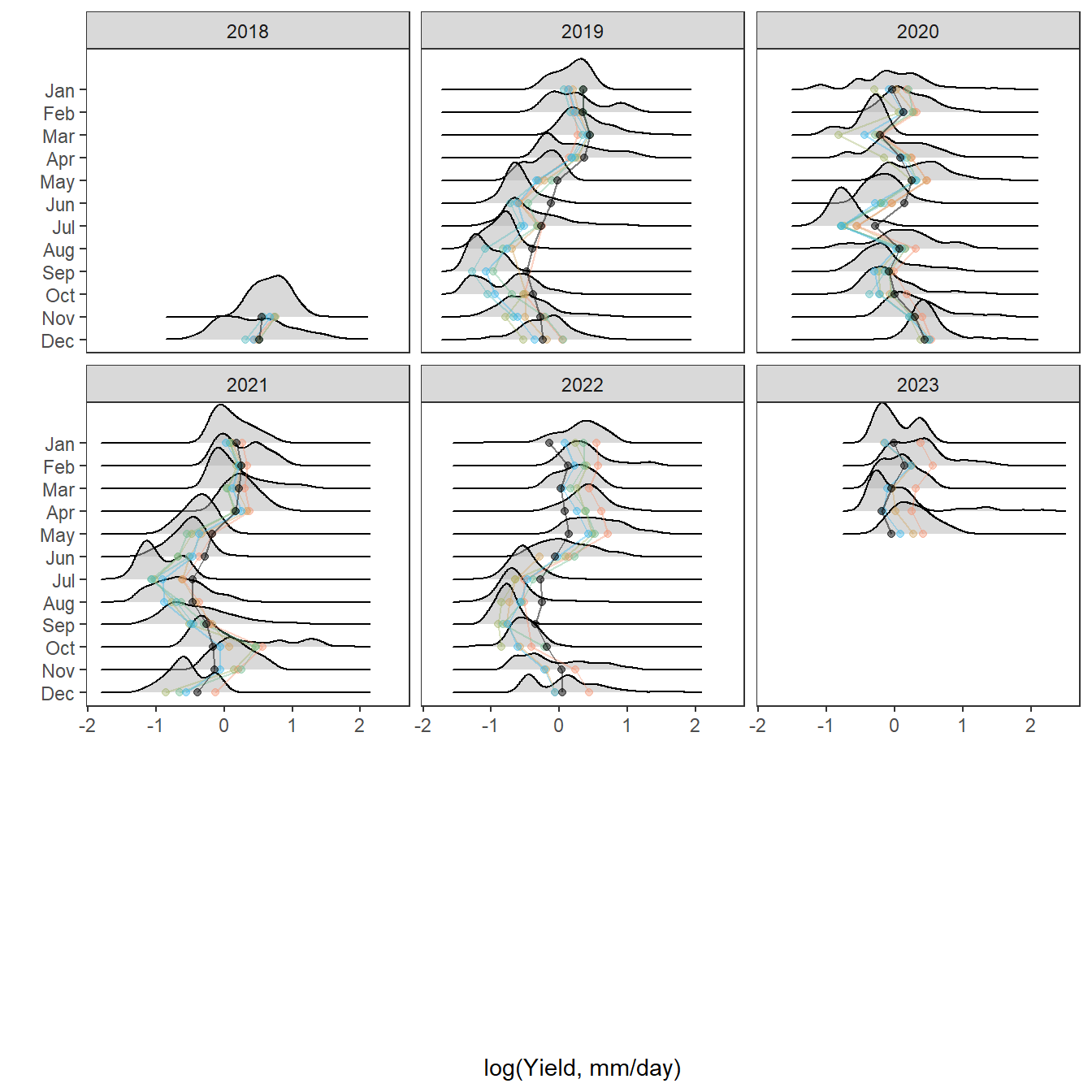
myridgesfun(bas = "Staunton River", orderr = stauntorder)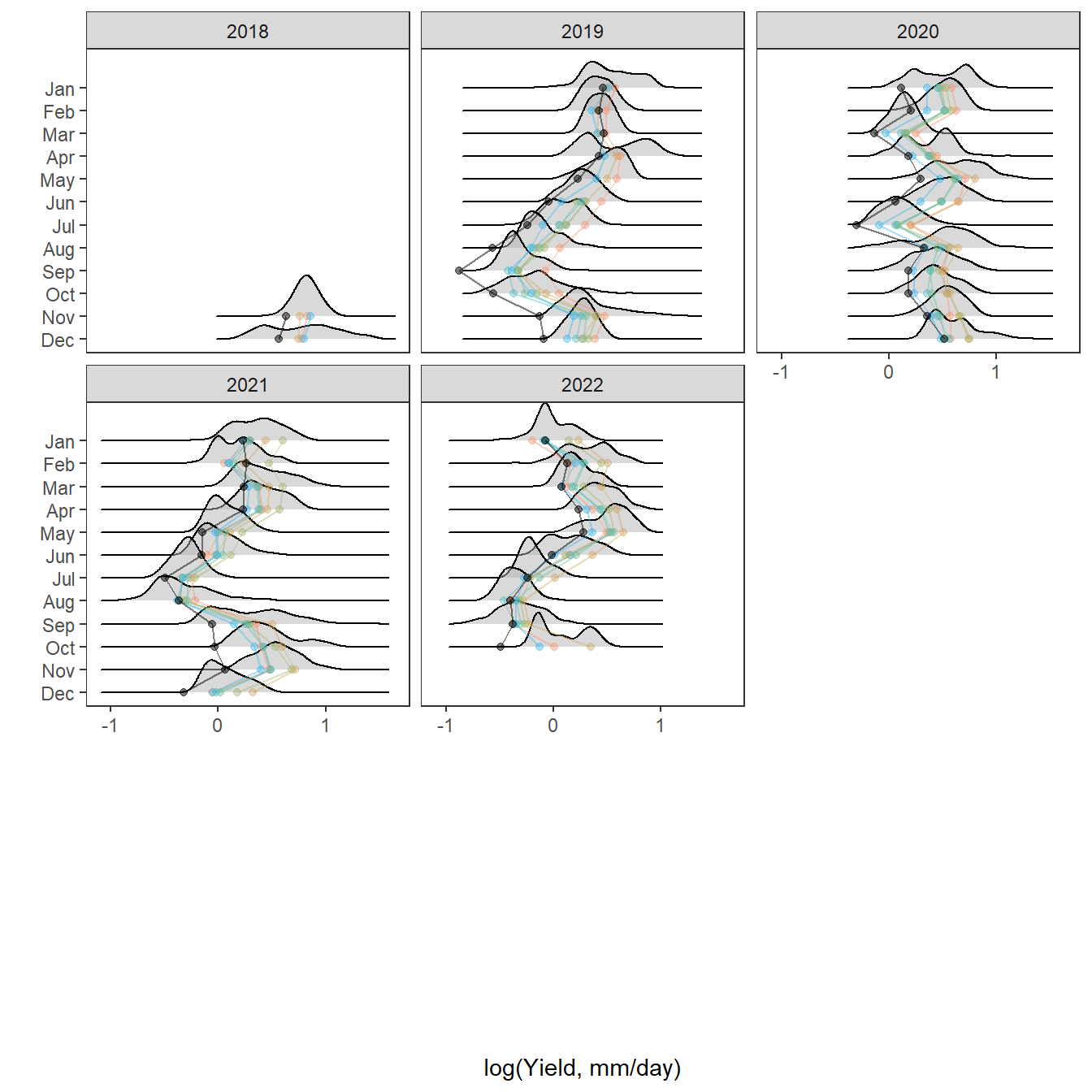
myridgesfun(bas = "Flathead", orderr = flatorder)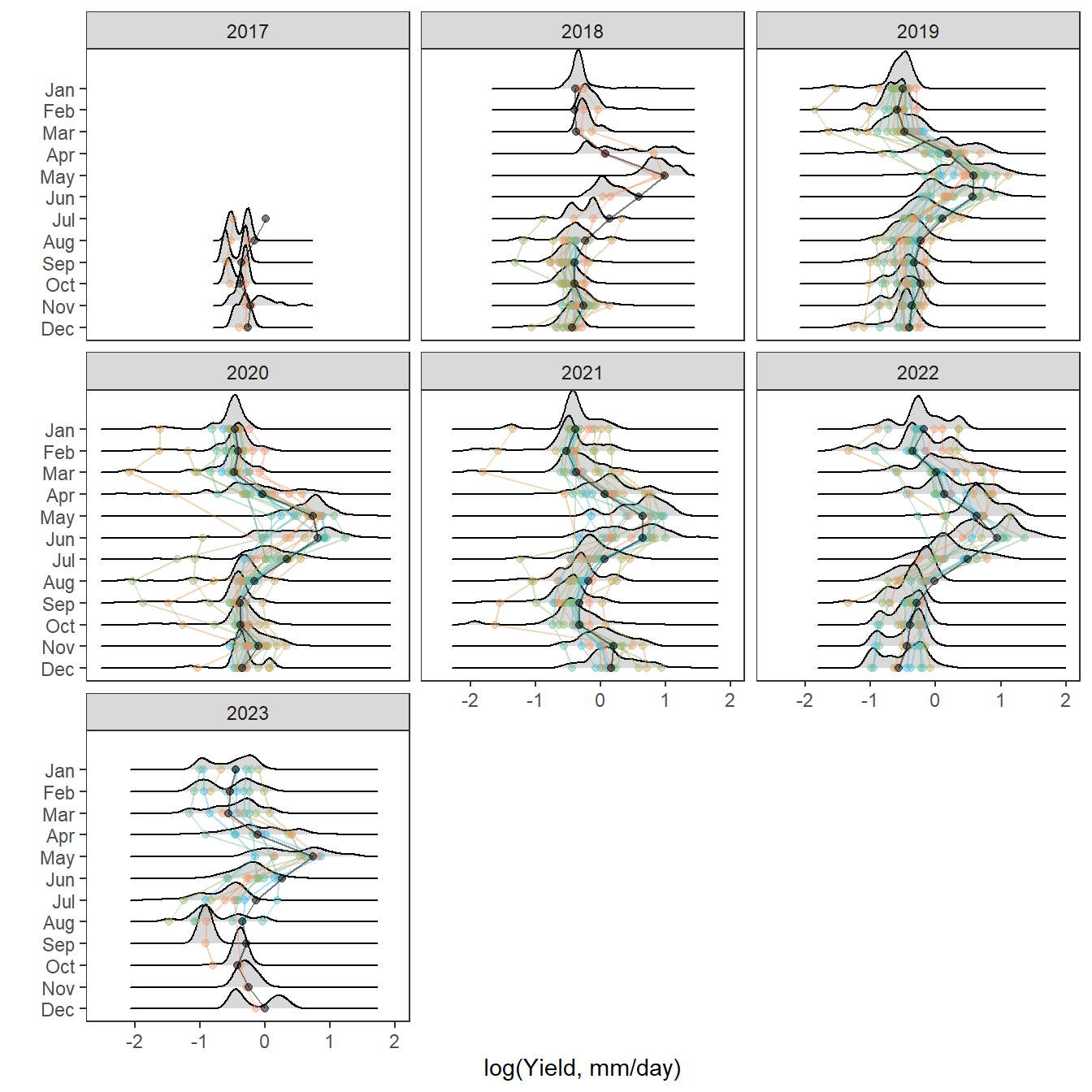
myridgesfun(bas = "Shields River", orderr = yellorder)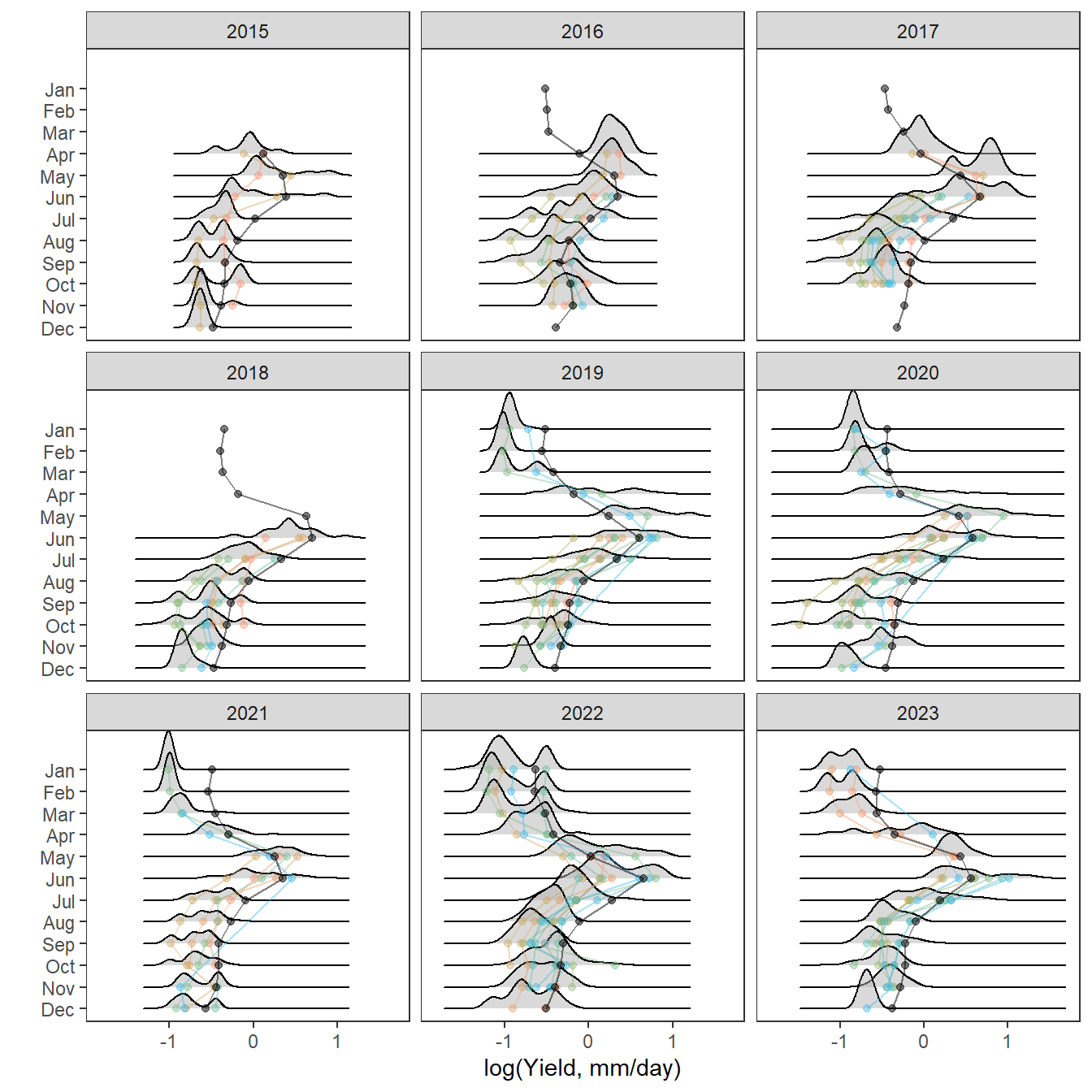
myridgesfun(bas = "Snake River", orderr = snakeorder)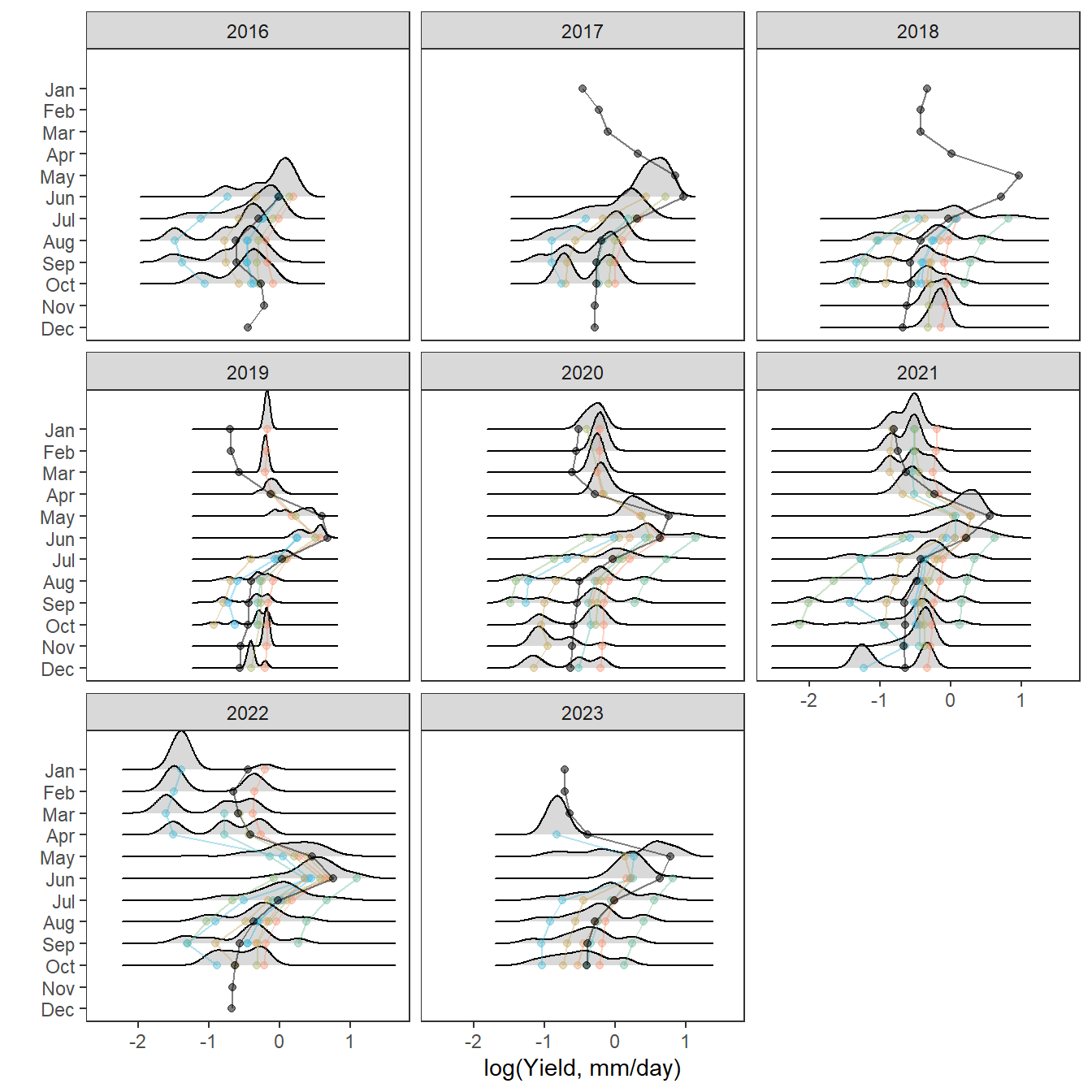
myridgesfun(bas = "Donner Blitzen", orderr = donnerorder)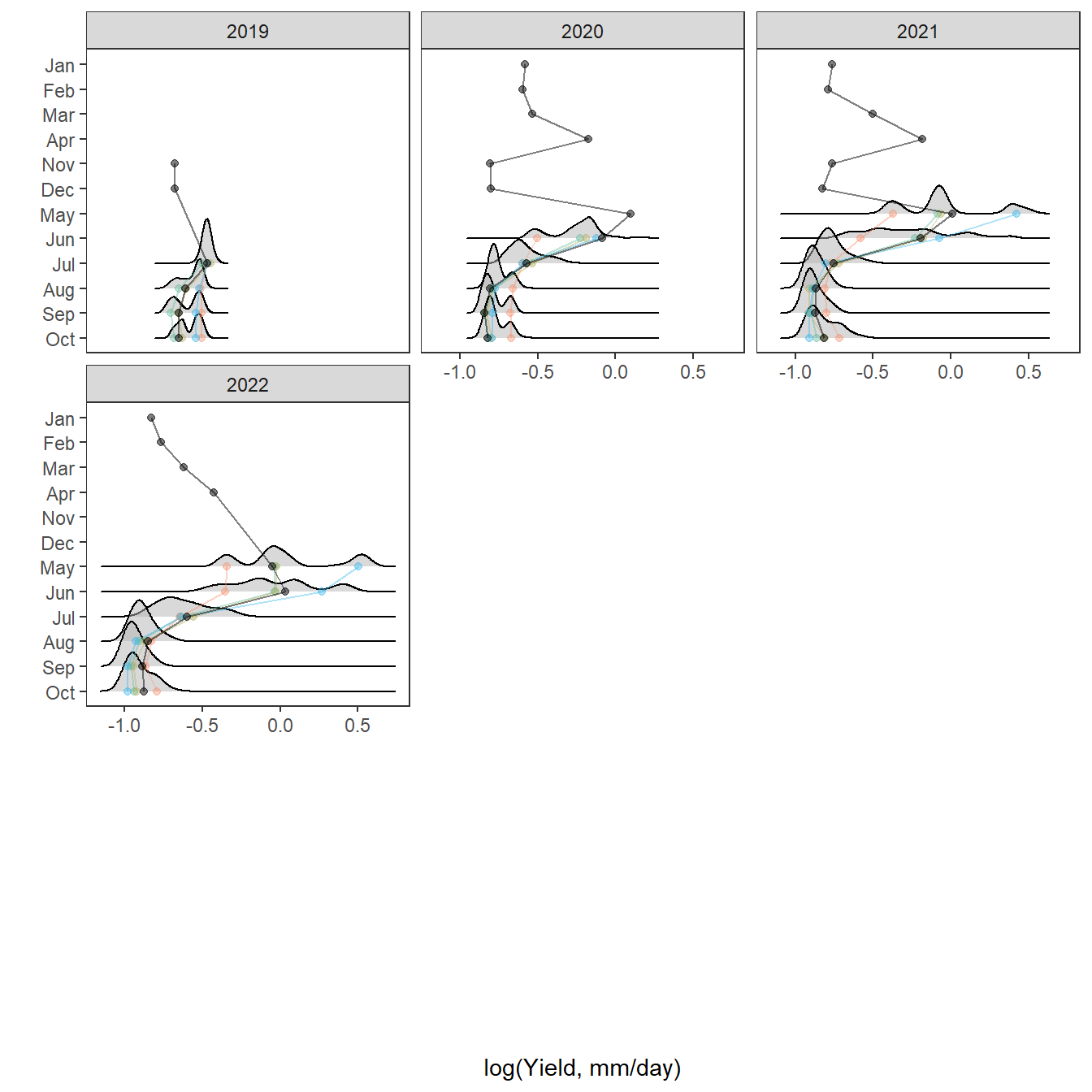
How repeatable is monthly mean flow among sites within basins? I.e., sites with higher than average yield always higher than average, or do lines cross frequently?
Calculate reputability (R) using the rptR package (Stoffel et al. 2017). Also see related papers by Nakagawa and Schielzeth. Repeatibility is calculated on mean monthly flow z-scored by month and year, which removes the effect of monthly and interannual variation in water availability. Results are very similar to models fit using raw yield data with big G flow as a fixed effect.
Generally, repeatibility appears to be greater for snowmelt-dominated basins, particularly in the summer. The “all data” plot is more difficult to interpret as we mostly have seasonal data in the snowmelt basins, particularly the Yellowstone and Snake. In actuality, I would expect the opposite pattern if we had complete time series data for all basins: low reputability for snowmelt basins and high repeatability for rainfall basins (for the same reasons as bow tie vs. wedge).
rp1 <- rpttib %>%
filter(basin != "Donner Blitzen", type == "zscore") %>%
mutate(basin = ifelse(basin == "Paine Run", "Paine",
ifelse(basin == "Staunton River", "Staunton",
ifelse(basin == "Snake River", "Snake",
ifelse(basin == "Shields River", "Yellowstone", basin))))) %>%
ggplot(aes(x = rboot, y = factor(basin, levels = rev(c("West Brook", "Staunton", "Paine", "Flathead", "Yellowstone", "Snake"))))) +
geom_density_ridges(quantile_lines = TRUE, quantiles = 2) +
xlim(0,1) +
theme_bw() + theme(legend.position = "none", panel.grid.major = element_blank(), panel.grid.minor = element_blank()) +
xlab("Bootstrapped repeatability (R)") + ylab("Basin") + ggtitle("All data") +
geom_text(data = rpttib %>% filter(basin != "Donner Blitzen", type == "zscore") %>%
mutate(basin = ifelse(basin == "Paine Run", "Paine",
ifelse(basin == "Staunton River", "Staunton",
ifelse(basin == "Snake River", "Snake",
ifelse(basin == "Shields River", "Yellowstone", basin)))))%>% group_by(basin) %>% summarise(rboot = median(rboot)),
aes(label = round(rboot, digits = 2)), position = position_nudge(y = -0.1), colour = "red", size=3.5)
rp2 <- rpttib_summer %>%
filter(basin != "Donner Blitzen", type == "zscore") %>%
mutate(basin = ifelse(basin == "Paine Run", "Paine",
ifelse(basin == "Staunton River", "Staunton",
ifelse(basin == "Snake River", "Snake",
ifelse(basin == "Shields River", "Yellowstone", basin))))) %>%
mutate(basin = factor(basin, levels = rev(c("West Brook", "Staunton", "Paine", "Flathead", "Yellowstone", "Snake")))) %>%
ggplot(aes(x = rboot, y = basin)) +
geom_density_ridges(quantile_lines = TRUE, quantiles = 2) +
# scale_fill_manual(values = rev(brewer.pal(6, "Set2"))) +
xlim(0,1) +
theme_bw() + theme(legend.position = "none", panel.grid.major = element_blank(), panel.grid.minor = element_blank()) +
xlab("Bootstrapped repeatability (R)") + ylab("Basin") + ggtitle("Summer only (JAS)") +
geom_text(data = rpttib_summer %>% filter(basin != "Donner Blitzen", type == "zscore") %>%
mutate(basin = ifelse(basin == "Paine Run", "Paine",
ifelse(basin == "Staunton River", "Staunton",
ifelse(basin == "Snake River", "Snake",
ifelse(basin == "Shields River", "Yellowstone", basin)))))%>% group_by(basin) %>% summarise(rboot = median(rboot)),
aes(label = round(rboot, digits = 2)), position = position_nudge(y = -0.1), colour = "red", size=3.5)
rp1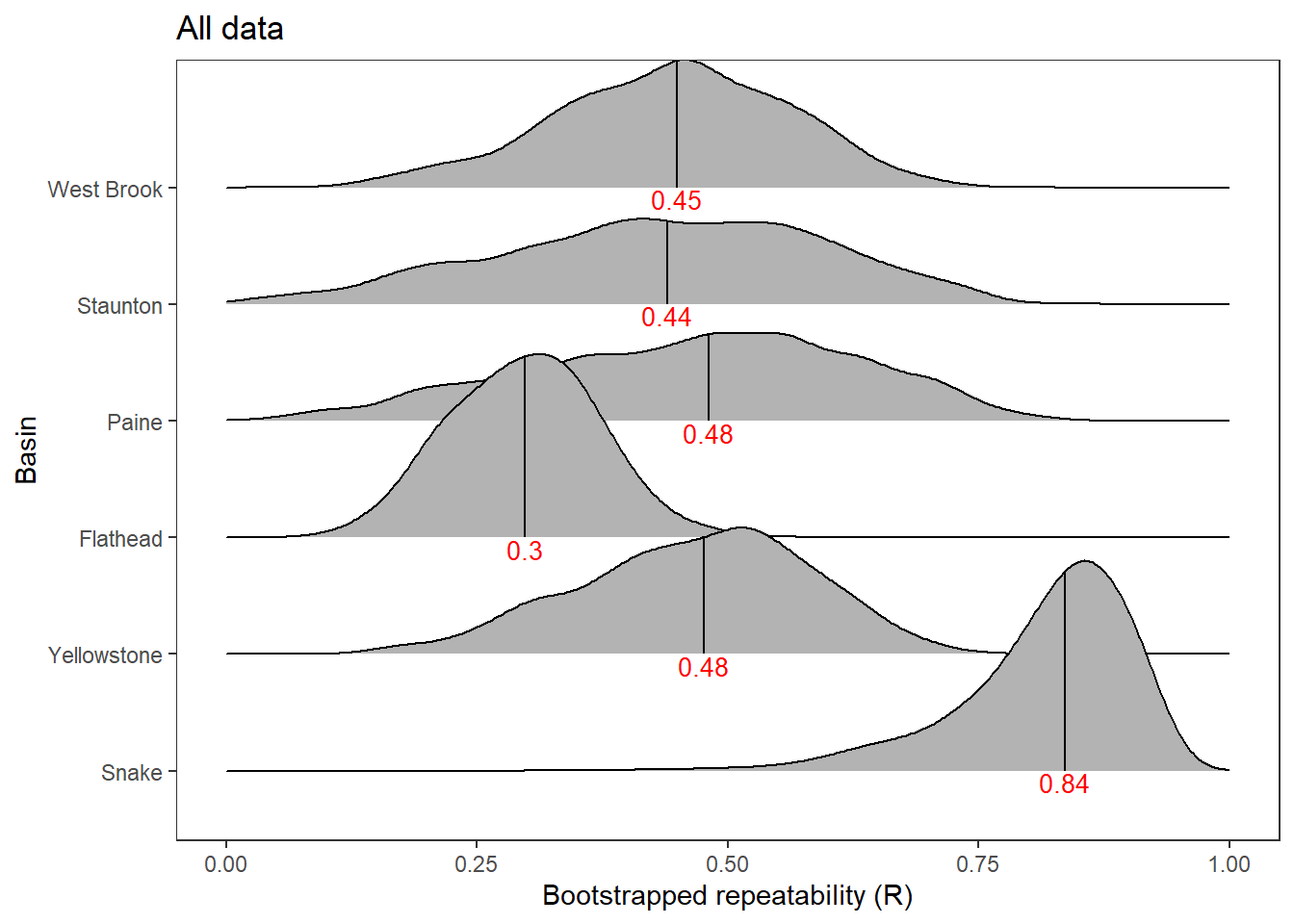
rp2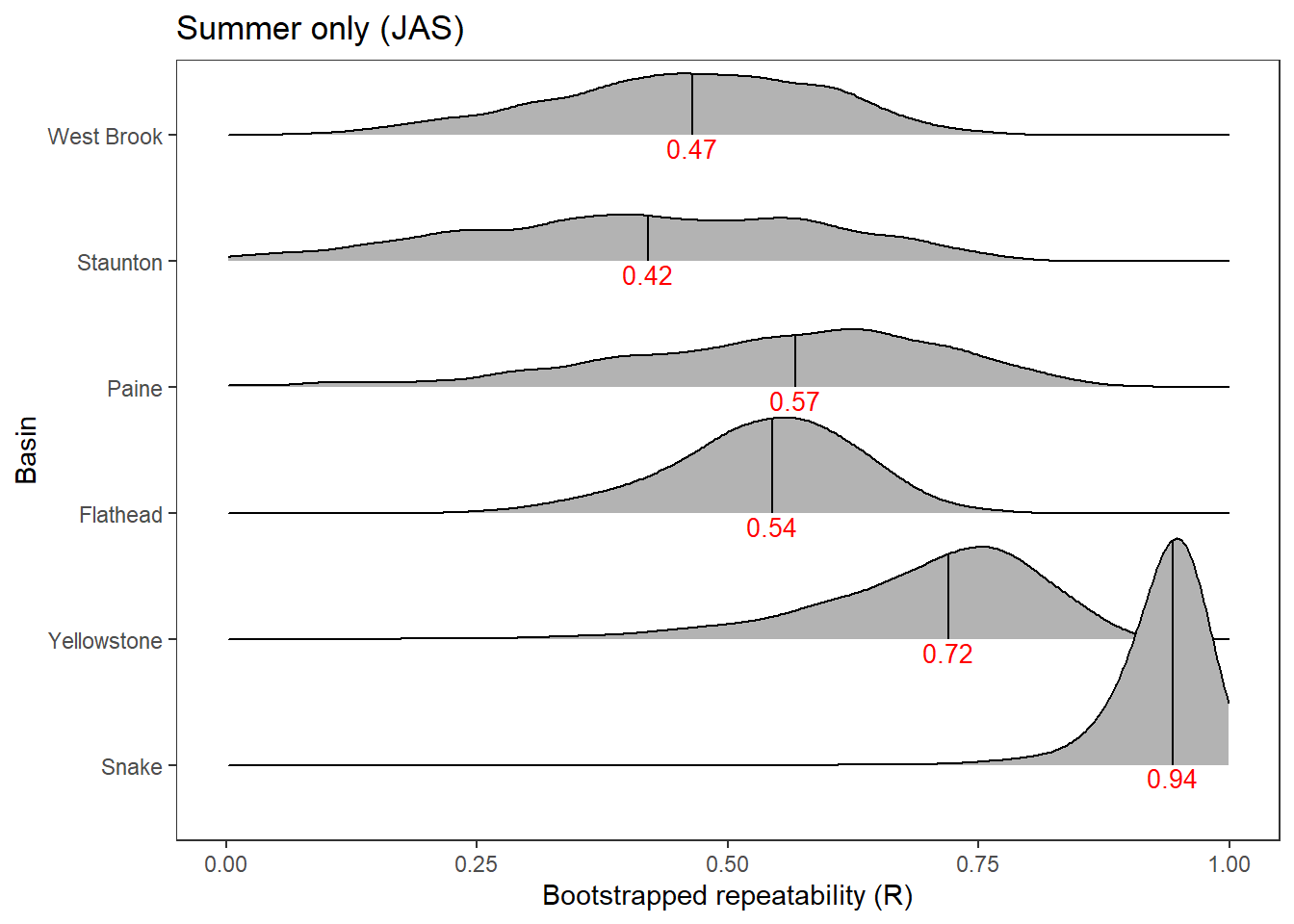
Write summer repeatability figure to file
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/MeanMonthlyFlow_SummerRepeatability.jpg", width = 4, height = 3.5, units = "in", res = 1000)
rpttib_summer %>%
filter(basin != "Donner Blitzen", type == "zscore") %>%
mutate(basin = ifelse(basin == "Paine Run", "Paine",
ifelse(basin == "Staunton River", "Staunton",
ifelse(basin == "Snake River", "Snake",
ifelse(basin == "Shields River", "Yellowstone", basin))))) %>%
mutate(basin = factor(basin, levels = rev(c("West Brook", "Staunton", "Paine", "Flathead", "Yellowstone", "Snake")))) %>%
ggplot(aes(x = rboot, y = basin)) +
geom_density_ridges(quantile_lines = TRUE, quantiles = 2, aes(fill = basin)) +
scale_fill_manual(values = rev(brewer.pal(6, "Set2"))) +
xlim(0,1) +
theme_bw() + theme(legend.position = "none", panel.grid.major = element_blank(), panel.grid.minor = element_blank(), axis.text = element_text(color = "black")) +
xlab("Bootstrapped repeatability (R)") + ylab("Basin") + #ggtitle("Summer only (JAS)") +
geom_text(data = rpttib_summer %>% filter(basin != "Donner Blitzen", type == "zscore") %>%
mutate(basin = ifelse(basin == "Paine Run", "Paine",
ifelse(basin == "Staunton River", "Staunton",
ifelse(basin == "Snake River", "Snake",
ifelse(basin == "Shields River", "Yellowstone", basin)))))%>% group_by(basin) %>% summarise(rboot = median(rboot)),
aes(label = round(rboot, digits = 2)), position = position_nudge(y = -0.15), colour = "black", size=3.5)
dev.off()png
2 myridgesfun2 <- function(bas, orderr, calyr) {
nsites <- length(orderr)
# daily data
td <- dat_clean %>% filter(basin == bas, CalendarYear == calyr) %>%
mutate(MonthName = factor(MonthName, levels = rev(c("Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sep", "Oct", "Nov", "Dec"))),
CalendarYear = factor(CalendarYear))
# summarize by month
td2 <- td %>%
group_by(subbasin, site_name, designation, CalendarYear, Month, MonthName) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
mutate(MonthName = factor(MonthName, levels = rev(c("Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sep", "Oct", "Nov", "Dec"))),
CalendarYear = factor(CalendarYear))
# big g data
tempdat_big <- dat_clean_big %>%
filter(basin == bas, date >= min(td$date), date <= max(td$date)) %>%
group_by(site_name, CalendarYear, Month, MonthName) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
mutate(MonthName = factor(MonthName, levels = rev(c("Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sep", "Oct", "Nov", "Dec"))),
CalendarYear = factor(CalendarYear))
return(ggplot(data = td2) +
geom_density_ridges(data = td, aes(x = logYield, y = MonthName), alpha = 0.5) +
geom_line(data = td2, aes(x = logYield, y = MonthName, group = site_name, color = factor(site_name, levels = orderr)), orientation = "y", alpha = 0.8) +
geom_point(data = td2, aes(x = logYield, y = MonthName, group = site_name, color = factor(site_name, levels = orderr)), alpha = 0.8) +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
geom_line(data = tempdat_big, aes(x = logYield, y = MonthName, group = site_name), orientation = "y", alpha = 0.5) +
geom_point(data = tempdat_big, aes(x = logYield, y = MonthName, group = site_name), alpha = 0.5) +
theme_bw() + theme(legend.position = "none", panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.title = element_blank(), axis.text = element_blank(), plot.title = element_text(size = 11)) +
xlim(-2.5,2)
#facet_wrap2(~CalendarYear, nrow = 3, ncol = 3, trim_blank = FALSE) +
#xlab("log(Yield, mm/day)") + ylab("")
)
}# arranged
ggarrange(p1, p2, ncol = 2, widths = c(1,0.7))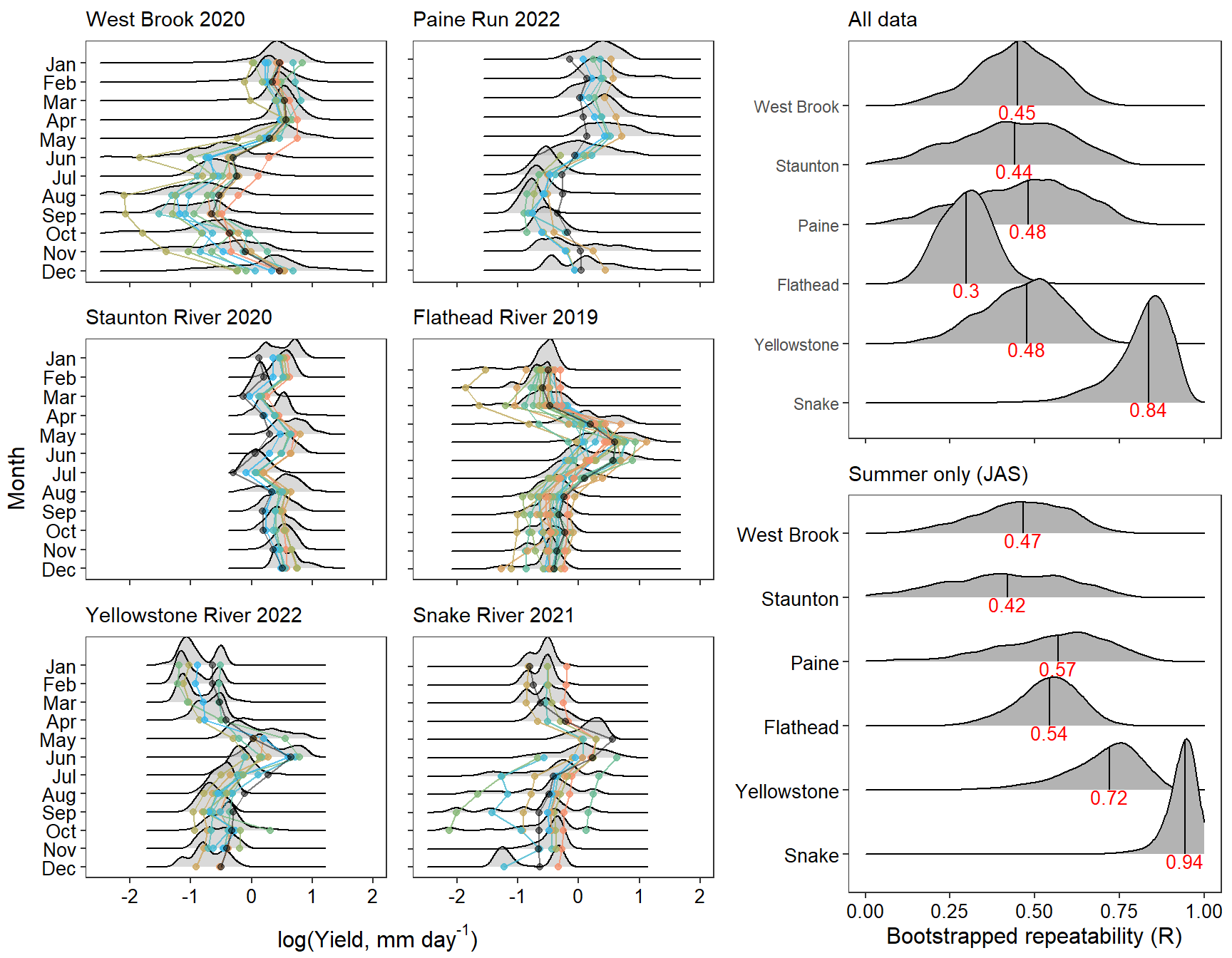
Summer exceedance curves and spatial variability in flow relative to (bootstrapped) temporal variability
### WEST BROOK
tempdat <- dat_clean %>%
filter(basin == "West Brook", !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = wborder))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "West Brook", date >= date("2020-01-01"), date <= date("2025-01-03"))
# exceedance
exceed_little <- tempdat %>%
group_by(site_name) %>%
arrange(desc(logYield), .by_group = TRUE) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield)) %>%
ungroup()
exceed_big <- tempdat_big %>%
arrange(desc(logYield), .by_group = TRUE) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield))
# plot
ewb1 <- ggplot() +
geom_line(data = exceed_little, aes(x = exceedance, y = logYield, color = factor(site_name, levels = wborder))) +
geom_line(data = exceed_big, aes(x = exceedance, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
#annotate("text", x = max(c(exceed_big$exceedance, exceed_little$exceedance), na.rm = TRUE), y = max(c(exceed_big$logYield, exceed_little$logYield), na.rm = TRUE), label = "The West Brook", hjust = 1, vjust = 1) +
scale_x_continuous(expand = c(0.02,0.02)) + scale_y_continuous(expand = c(0.02,0.02)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
# summarize
exceed_little_sd <- exceed_little %>%
mutate(exceedance = round(exceedance, digits = 0)) %>%
group_by(site_name, exceedance) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
group_by(exceedance) %>%
summarize(sdyield = sd(logYield)) %>%
ungroup()
exceed_big_sum <- exceed_big %>%
mutate(exceedance = round(exceedance, digits = 0)) %>%
group_by(exceedance) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
filter(!is.na(logYield))
# bootstrap
nreps <- 1000
randsampsd <- c()
for (i in 1:nreps) { randsampsd[i] <- sd(sample(exceed_big_sum$logYield, size = nsites, replace = FALSE)) }
exceed_little_sd_exp <- do.call("rbind", replicate(nreps, exceed_little_sd, simplify = FALSE))
exceed_little_sd_exp <- exceed_little_sd_exp %>%
mutate(sdbigtemp = rep(randsampsd, each = nrow(exceed_little_sd)),
replicate = rep(c(1:nreps), each = nrow(exceed_little_sd)),
reldiff = ((sdyield-sdbigtemp)/sdbigtemp)*100)
exceed_little_sd_exp_sum <- exceed_little_sd_exp %>%
group_by(exceedance) %>%
summarize(q025 = quantile(reldiff, probs = 0.025),
q10 = quantile(reldiff, probs = 0.10),
q50 = quantile(reldiff, probs = 0.5),
q90 = quantile(reldiff, probs = 0.90),
q975 = quantile(reldiff, probs = 0.975))
# plot
vwb1 <- exceed_little_sd_exp_sum %>%
ggplot() +
geom_ribbon(aes(x = exceedance, ymin = q025, ymax = q975), fill = "grey85") +
geom_ribbon(aes(x = exceedance, ymin = q10, ymax = q90), fill = "grey70") +
geom_line(aes(x = exceedance, y = q50), color = "black", size = 1) +
geom_abline(intercept = 0, slope = 0, linetype = "dashed") +
scale_x_continuous(expand = c(0.02,0.02)) + scale_y_continuous(expand = c(0.02,0.02)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### PAINE RUN
tempdat <- dat_clean %>%
filter(basin == "Paine Run", !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = paineorder))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Paine Run", date >= as_date("2018-11-07"), date <= as_date("2023-05-15"))
# exceedance
exceed_little <- tempdat %>%
group_by(site_name) %>%
arrange(desc(logYield), .by_group = TRUE) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield)) %>%
ungroup()
exceed_big <- tempdat_big %>%
arrange(desc(logYield), .by_group = TRUE) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield))
# plot
epa1 <- ggplot() +
geom_line(data = exceed_little, aes(x = exceedance, y = logYield, color = factor(site_name, levels = paineorder))) +
geom_line(data = exceed_big, aes(x = exceedance, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
#annotate("text", x = max(c(exceed_big$exceedance, exceed_little$exceedance), na.rm = TRUE), y = max(c(exceed_big$logYield, exceed_little$logYield), na.rm = TRUE), label = "Paine Run", hjust = 1, vjust = 1) +
scale_x_continuous(expand = c(0.02,0.02)) + scale_y_continuous(expand = c(0.02,0.02)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
# summarize
exceed_little_sd <- exceed_little %>%
mutate(exceedance = round(exceedance, digits = 0)) %>%
group_by(site_name, exceedance) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
group_by(exceedance) %>%
summarize(sdyield = sd(logYield)) %>%
ungroup()
exceed_big_sum <- exceed_big %>%
mutate(exceedance = round(exceedance, digits = 0)) %>%
group_by(exceedance) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
filter(!is.na(logYield))
# bootstrap
nreps <- 1000
randsampsd <- c()
for (i in 1:nreps) { randsampsd[i] <- sd(sample(exceed_big_sum$logYield, size = nsites, replace = FALSE)) }
exceed_little_sd_exp <- do.call("rbind", replicate(nreps, exceed_little_sd, simplify = FALSE))
exceed_little_sd_exp <- exceed_little_sd_exp %>%
mutate(sdbigtemp = rep(randsampsd, each = nrow(exceed_little_sd)),
replicate = rep(c(1:nreps), each = nrow(exceed_little_sd)),
reldiff = ((sdyield-sdbigtemp)/sdbigtemp)*100)
exceed_little_sd_exp_sum <- exceed_little_sd_exp %>%
group_by(exceedance) %>%
summarize(q025 = quantile(reldiff, probs = 0.025),
q10 = quantile(reldiff, probs = 0.10),
q50 = quantile(reldiff, probs = 0.5),
q90 = quantile(reldiff, probs = 0.90),
q975 = quantile(reldiff, probs = 0.975))
# plot
vpa1 <- exceed_little_sd_exp_sum %>%
ggplot() +
geom_ribbon(aes(x = exceedance, ymin = q025, ymax = q975), fill = "grey85") +
geom_ribbon(aes(x = exceedance, ymin = q10, ymax = q90), fill = "grey70") +
geom_line(aes(x = exceedance, y = q50), color = "black", size = 1) +
geom_abline(intercept = 0, slope = 0, linetype = "dashed") +
scale_x_continuous(expand = c(0.02,0.02)) + scale_y_continuous(expand = c(0.02,0.02)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### STAUNTON RIVER
tempdat <- dat_clean %>%
filter(basin == "Staunton River", !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = stauntorder))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Staunton River", date >= as_date("2018-11-07"), date <= as_date("2022-10-19"))
# exceedance
exceed_little <- tempdat %>%
group_by(site_name) %>%
arrange(desc(logYield), .by_group = TRUE) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield)) %>%
ungroup()
exceed_big <- tempdat_big %>%
arrange(desc(logYield), .by_group = TRUE) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield))
# plot
est1 <- ggplot() +
geom_line(data = exceed_little, aes(x = exceedance, y = logYield, color = factor(site_name, levels = stauntorder))) +
geom_line(data = exceed_big, aes(x = exceedance, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
#annotate("text", x = max(c(exceed_big$exceedance, exceed_little$exceedance), na.rm = TRUE), y = max(c(exceed_big$logYield, exceed_little$logYield), na.rm = TRUE), label = "Staunton River", hjust = 1, vjust = 1) +
scale_x_continuous(expand = c(0.02,0.02)) + scale_y_continuous(expand = c(0.02,0.02)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
# summarize
exceed_little_sd <- exceed_little %>%
mutate(exceedance = round(exceedance, digits = 0)) %>%
group_by(site_name, exceedance) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
group_by(exceedance) %>%
summarize(sdyield = sd(logYield)) %>%
ungroup()
exceed_big_sum <- exceed_big %>%
mutate(exceedance = round(exceedance, digits = 0)) %>%
group_by(exceedance) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
filter(!is.na(logYield))
# bootstrap
nreps <- 1000
randsampsd <- c()
for (i in 1:nreps) { randsampsd[i] <- sd(sample(exceed_big_sum$logYield, size = nsites, replace = FALSE)) }
exceed_little_sd_exp <- do.call("rbind", replicate(nreps, exceed_little_sd, simplify = FALSE))
exceed_little_sd_exp <- exceed_little_sd_exp %>%
mutate(sdbigtemp = rep(randsampsd, each = nrow(exceed_little_sd)),
replicate = rep(c(1:nreps), each = nrow(exceed_little_sd)),
reldiff = ((sdyield-sdbigtemp)/sdbigtemp)*100)
exceed_little_sd_exp_sum <- exceed_little_sd_exp %>%
group_by(exceedance) %>%
summarize(q025 = quantile(reldiff, probs = 0.025),
q10 = quantile(reldiff, probs = 0.10),
q50 = quantile(reldiff, probs = 0.5),
q90 = quantile(reldiff, probs = 0.90),
q975 = quantile(reldiff, probs = 0.975))
# plot
vst1 <- exceed_little_sd_exp_sum %>%
ggplot() +
geom_ribbon(aes(x = exceedance, ymin = q025, ymax = q975), fill = "grey85") +
geom_ribbon(aes(x = exceedance, ymin = q10, ymax = q90), fill = "grey70") +
geom_line(aes(x = exceedance, y = q50), color = "black", size = 1) +
geom_abline(intercept = 0, slope = 0, linetype = "dashed") +
scale_x_continuous(expand = c(0.02,0.02)) + scale_y_continuous(expand = c(0.02,0.02)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### FLATHEAD
tempdat <- dat_clean %>%
filter(basin == "Flathead", date >= date("2017-07-29"), date <= date("2023-12-11"), !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = flatorder))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Flathead", date >= date("2017-07-29"), date <= date("2023-12-11"))
# exceedance
exceed_little <- tempdat %>%
group_by(site_name) %>%
arrange(desc(logYield), .by_group = TRUE) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield)) %>%
ungroup()
exceed_big <- tempdat_big %>%
arrange(desc(logYield), .by_group = TRUE) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield))
# plot
efl1 <- ggplot() +
geom_line(data = exceed_little, aes(x = exceedance, y = logYield, color = factor(site_name, levels = flatorder))) +
geom_line(data = exceed_big, aes(x = exceedance, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
#annotate("text", x = max(c(exceed_big$exceedance, exceed_little$exceedance), na.rm = TRUE), y = max(c(exceed_big$logYield, exceed_little$logYield), na.rm = TRUE), label = "Flathead River", hjust = 1, vjust = 1) +
scale_x_continuous(expand = c(0.02,0.02)) + scale_y_continuous(expand = c(0.02,0.02)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
# summarize
exceed_little_sd <- exceed_little %>%
mutate(exceedance = round(exceedance, digits = 0)) %>%
group_by(site_name, exceedance) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
group_by(exceedance) %>%
summarize(sdyield = sd(logYield)) %>%
ungroup()
exceed_big_sum <- exceed_big %>%
mutate(exceedance = round(exceedance, digits = 0)) %>%
group_by(exceedance) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
filter(!is.na(logYield))
# bootstrap
nreps <- 1000
randsampsd <- c()
for (i in 1:nreps) { randsampsd[i] <- sd(sample(exceed_big_sum$logYield, size = nsites, replace = FALSE)) }
exceed_little_sd_exp <- do.call("rbind", replicate(nreps, exceed_little_sd, simplify = FALSE))
exceed_little_sd_exp <- exceed_little_sd_exp %>%
mutate(sdbigtemp = rep(randsampsd, each = nrow(exceed_little_sd)),
replicate = rep(c(1:nreps), each = nrow(exceed_little_sd)),
reldiff = ((sdyield-sdbigtemp)/sdbigtemp)*100)
exceed_little_sd_exp_sum <- exceed_little_sd_exp %>%
group_by(exceedance) %>%
summarize(q025 = quantile(reldiff, probs = 0.025),
q10 = quantile(reldiff, probs = 0.10),
q50 = quantile(reldiff, probs = 0.5),
q90 = quantile(reldiff, probs = 0.90),
q975 = quantile(reldiff, probs = 0.975))
# plot
vfl1 <- exceed_little_sd_exp_sum %>%
ggplot() +
geom_ribbon(aes(x = exceedance, ymin = q025, ymax = q975), fill = "grey85") +
geom_ribbon(aes(x = exceedance, ymin = q10, ymax = q90), fill = "grey70") +
geom_line(aes(x = exceedance, y = q50), color = "black", size = 1) +
geom_abline(intercept = 0, slope = 0, linetype = "dashed") +
scale_x_continuous(expand = c(0.02,0.02)) + scale_y_continuous(expand = c(0.02,0.02)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### YELLOWSTONE
tempdat <- dat_clean %>%
filter(basin == "Shields River", !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = yellorder))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Shields River", date >= date("2015-04-01"), date <= date("2023-12-31"))
# exceedance
exceed_little <- tempdat %>%
group_by(site_name) %>%
arrange(desc(logYield), .by_group = TRUE) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield)) %>%
ungroup()
exceed_big <- tempdat_big %>%
arrange(desc(logYield), .by_group = TRUE) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield))
# plot
eye1 <- ggplot() +
geom_line(data = exceed_little, aes(x = exceedance, y = logYield, color = factor(site_name, levels = yellorder))) +
geom_line(data = exceed_big, aes(x = exceedance, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
#annotate("text", x = max(c(exceed_big$exceedance, exceed_little$exceedance), na.rm = TRUE), y = max(c(exceed_big$logYield, exceed_little$logYield), na.rm = TRUE), label = "Yellowstone River", hjust = 1, vjust = 1) +
scale_x_continuous(expand = c(0.02,0.02)) + scale_y_continuous(expand = c(0.02,0.02)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
# summarize
exceed_little_sd <- exceed_little %>%
mutate(exceedance = round(exceedance, digits = 0)) %>%
group_by(site_name, exceedance) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
group_by(exceedance) %>%
summarize(sdyield = sd(logYield)) %>%
ungroup()
exceed_big_sum <- exceed_big %>%
mutate(exceedance = round(exceedance, digits = 0)) %>%
group_by(exceedance) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
filter(!is.na(logYield))
# bootstrap
nreps <- 1000
randsampsd <- c()
for (i in 1:nreps) { randsampsd[i] <- sd(sample(exceed_big_sum$logYield, size = nsites, replace = FALSE)) }
exceed_little_sd_exp <- do.call("rbind", replicate(nreps, exceed_little_sd, simplify = FALSE))
exceed_little_sd_exp <- exceed_little_sd_exp %>%
mutate(sdbigtemp = rep(randsampsd, each = nrow(exceed_little_sd)),
replicate = rep(c(1:nreps), each = nrow(exceed_little_sd)),
reldiff = ((sdyield-sdbigtemp)/sdbigtemp)*100)
exceed_little_sd_exp_sum <- exceed_little_sd_exp %>%
group_by(exceedance) %>%
summarize(q025 = quantile(reldiff, probs = 0.025),
q10 = quantile(reldiff, probs = 0.10),
q50 = quantile(reldiff, probs = 0.5),
q90 = quantile(reldiff, probs = 0.90),
q975 = quantile(reldiff, probs = 0.975))
# plot
vye1 <- exceed_little_sd_exp_sum %>%
ggplot() +
geom_ribbon(aes(x = exceedance, ymin = q025, ymax = q975), fill = "grey85") +
geom_ribbon(aes(x = exceedance, ymin = q10, ymax = q90), fill = "grey70") +
geom_line(aes(x = exceedance, y = q50), color = "black", size = 1) +
geom_abline(intercept = 0, slope = 0, linetype = "dashed") +
scale_x_continuous(expand = c(0.02,0.02)) + scale_y_continuous(expand = c(0.02,0.02)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### SNAKE
tempdat <- dat_clean %>%
filter(basin == "Snake River", !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = snakeorder))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Snake River", date >= date("2016-04-01"), date <= date("2023-10-03"))
# exceedance
exceed_little <- tempdat %>%
group_by(site_name) %>%
arrange(desc(logYield), .by_group = TRUE) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield)) %>%
ungroup()
exceed_big <- tempdat_big %>%
arrange(desc(logYield), .by_group = TRUE) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield))
# plot
esn1 <- ggplot() +
geom_line(data = exceed_little, aes(x = exceedance, y = logYield, color = factor(site_name, levels = snakeorder))) +
geom_line(data = exceed_big, aes(x = exceedance, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
#annotate("text", x = max(c(exceed_big$exceedance, exceed_little$exceedance), na.rm = TRUE), y = max(c(exceed_big$logYield, exceed_little$logYield), na.rm = TRUE), label = "Snake River", hjust = 1, vjust = 1) +
scale_x_continuous(expand = c(0.02,0.02)) + scale_y_continuous(expand = c(0.02,0.02)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
# summarize
exceed_little_sd <- exceed_little %>%
mutate(exceedance = round(exceedance, digits = 0)) %>%
group_by(site_name, exceedance) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
group_by(exceedance) %>%
summarize(sdyield = sd(logYield)) %>%
ungroup()
exceed_big_sum <- exceed_big %>%
mutate(exceedance = round(exceedance, digits = 0)) %>%
group_by(exceedance) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
filter(!is.na(logYield))
# bootstrap
nreps <- 1000
randsampsd <- c()
for (i in 1:nreps) { randsampsd[i] <- sd(sample(exceed_big_sum$logYield, size = nsites, replace = FALSE)) }
exceed_little_sd_exp <- do.call("rbind", replicate(nreps, exceed_little_sd, simplify = FALSE))
exceed_little_sd_exp <- exceed_little_sd_exp %>%
mutate(sdbigtemp = rep(randsampsd, each = nrow(exceed_little_sd)),
replicate = rep(c(1:nreps), each = nrow(exceed_little_sd)),
reldiff = ((sdyield-sdbigtemp)/sdbigtemp)*100)
exceed_little_sd_exp_sum <- exceed_little_sd_exp %>%
group_by(exceedance) %>%
summarize(q025 = quantile(reldiff, probs = 0.025),
q10 = quantile(reldiff, probs = 0.10),
q50 = quantile(reldiff, probs = 0.5),
q90 = quantile(reldiff, probs = 0.90),
q975 = quantile(reldiff, probs = 0.975))
# plot
vsn1 <- exceed_little_sd_exp_sum %>%
ggplot() +
geom_ribbon(aes(x = exceedance, ymin = q025, ymax = q975), fill = "grey85") +
geom_ribbon(aes(x = exceedance, ymin = q10, ymax = q90), fill = "grey70") +
geom_line(aes(x = exceedance, y = q50), color = "black", size = 1) +
geom_abline(intercept = 0, slope = 0, linetype = "dashed") +
scale_x_continuous(expand = c(0.02,0.02)) + scale_y_continuous(expand = c(0.02,0.02)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
### DONNER BLITZEN
tempdat <- dat_clean %>%
filter(basin == "Donner Blitzen", !is.na(logYield), Month %in% c(7:9)) %>%
mutate(site_name = factor(site_name, levels = donnerorder))
nsites <- length(unique(tempdat$site_name))
tempdat_big <- dat_clean_big %>% filter(basin == "Donner Blitzen", date >= date("2019-06-01"), date <= as_date("2022-12-31"))
# exceedance
exceed_little <- tempdat %>%
group_by(site_name) %>%
arrange(desc(logYield), .by_group = TRUE) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield)) %>%
ungroup()
exceed_big <- tempdat_big %>%
arrange(desc(logYield), .by_group = TRUE) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield))
# plot
edb1 <- ggplot() +
geom_line(data = exceed_little, aes(x = exceedance, y = logYield, color = factor(site_name, levels = donnerorder))) +
geom_line(data = exceed_big, aes(x = exceedance, y = logYield), color = "grey40") +
scale_color_manual(values = cet_pal(nsites, name = "i1")) +
#annotate("text", x = max(c(exceed_big$exceedance, exceed_little$exceedance), na.rm = TRUE), y = max(c(exceed_big$logYield, exceed_little$logYield), na.rm = TRUE), label = "Donner und Blitzen River", hjust = 1, vjust = 1) +
scale_x_continuous(expand = c(0.02,0.02)) + scale_y_continuous(expand = c(0.02,0.02)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))
# summarize
exceed_little_sd <- exceed_little %>%
mutate(exceedance = round(exceedance, digits = 0)) %>%
group_by(site_name, exceedance) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
group_by(exceedance) %>%
summarize(sdyield = sd(logYield)) %>%
ungroup()
exceed_big_sum <- exceed_big %>%
mutate(exceedance = round(exceedance, digits = 0)) %>%
group_by(exceedance) %>%
summarize(logYield = mean(logYield)) %>%
ungroup() %>%
filter(!is.na(logYield))
# bootstrap
nreps <- 1000
randsampsd <- c()
for (i in 1:nreps) { randsampsd[i] <- sd(sample(exceed_big_sum$logYield, size = nsites, replace = FALSE)) }
exceed_little_sd_exp <- do.call("rbind", replicate(nreps, exceed_little_sd, simplify = FALSE))
exceed_little_sd_exp <- exceed_little_sd_exp %>%
mutate(sdbigtemp = rep(randsampsd, each = nrow(exceed_little_sd)),
replicate = rep(c(1:nreps), each = nrow(exceed_little_sd)),
reldiff = ((sdyield-sdbigtemp)/sdbigtemp)*100)
exceed_little_sd_exp_sum <- exceed_little_sd_exp %>%
group_by(exceedance) %>%
summarize(q025 = quantile(reldiff, probs = 0.025),
q10 = quantile(reldiff, probs = 0.10),
q50 = quantile(reldiff, probs = 0.5),
q90 = quantile(reldiff, probs = 0.90),
q975 = quantile(reldiff, probs = 0.975))
# plot
vdb1 <- exceed_little_sd_exp_sum %>%
ggplot() +
geom_ribbon(aes(x = exceedance, ymin = q025, ymax = q975), fill = "grey85") +
geom_ribbon(aes(x = exceedance, ymin = q10, ymax = q90), fill = "grey70") +
geom_line(aes(x = exceedance, y = q50), color = "black", size = 1) +
geom_abline(intercept = 0, slope = 0, linetype = "dashed") +
scale_x_continuous(expand = c(0.02,0.02)) + scale_y_continuous(expand = c(0.02,0.02)) +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none",
axis.title = element_blank(), plot.margin = margin(t = 1, r = 5, b = 2, l = 2), axis.text = element_text(color = "black"))Generate plot
annotate_figure(ggarrange(annotate_figure(egg::ggarrange(ewb1, epa1, est1, efl1, eye1, esn1, edb1, ncol = 1), left = "log(Yield, mm/day)"),
annotate_figure(egg::ggarrange(vwb1, vpa1, vst1, vfl1, vye1, vsn1, vdb1, ncol = 1), left = "Relative spatial variation in flow (%)"),
ncol = 2), bottom = "Exceedance probability")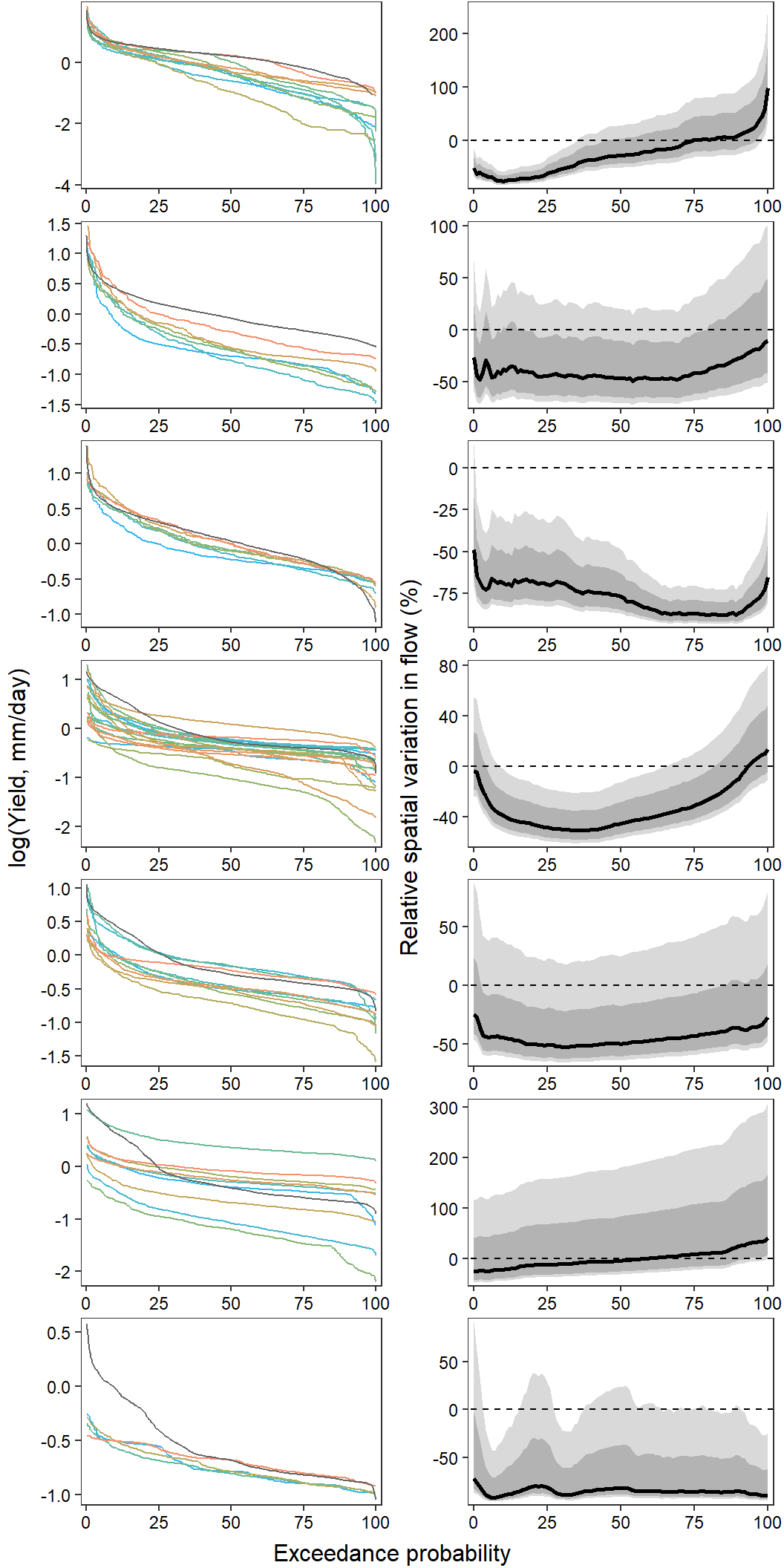
Write to file
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/EcoD_exceedance_new.jpg", width = 5.5, height = 11, units = "in", res = 1000)
annotate_figure(ggarrange(annotate_figure(egg::ggarrange(ewb1, epa1, est1, efl1, eye1, esn1, edb1, ncol = 1), left = "log(Yield, mm/day)"),
annotate_figure(egg::ggarrange(vwb1, vpa1, vst1, vfl1, vye1, vsn1, vdb1, ncol = 1), left = "Relative spatial variation in flow (%)"),
ncol = 2), bottom = "Exceedance probability")
dev.off()png
2 p <- ggarrange(annotate_figure(egg::ggarrange(pwb1, ppa1, pst1, pfl1, pye1, psn1, pdb1, ncol = 1),
bottom = text_grob("Date\n ", size = 11), left = text_grob("log(Yield, mm/day)", size = 11, rot = 90)),
annotate_figure(egg::ggarrange(ewb1, epa1, est1, efl1, eye1, esn1, edb1, ncol = 1),
bottom = text_grob("Exceedance probability \n(summer only)", size = 11)),
ncol = 2, widths = c(1,0.4))
p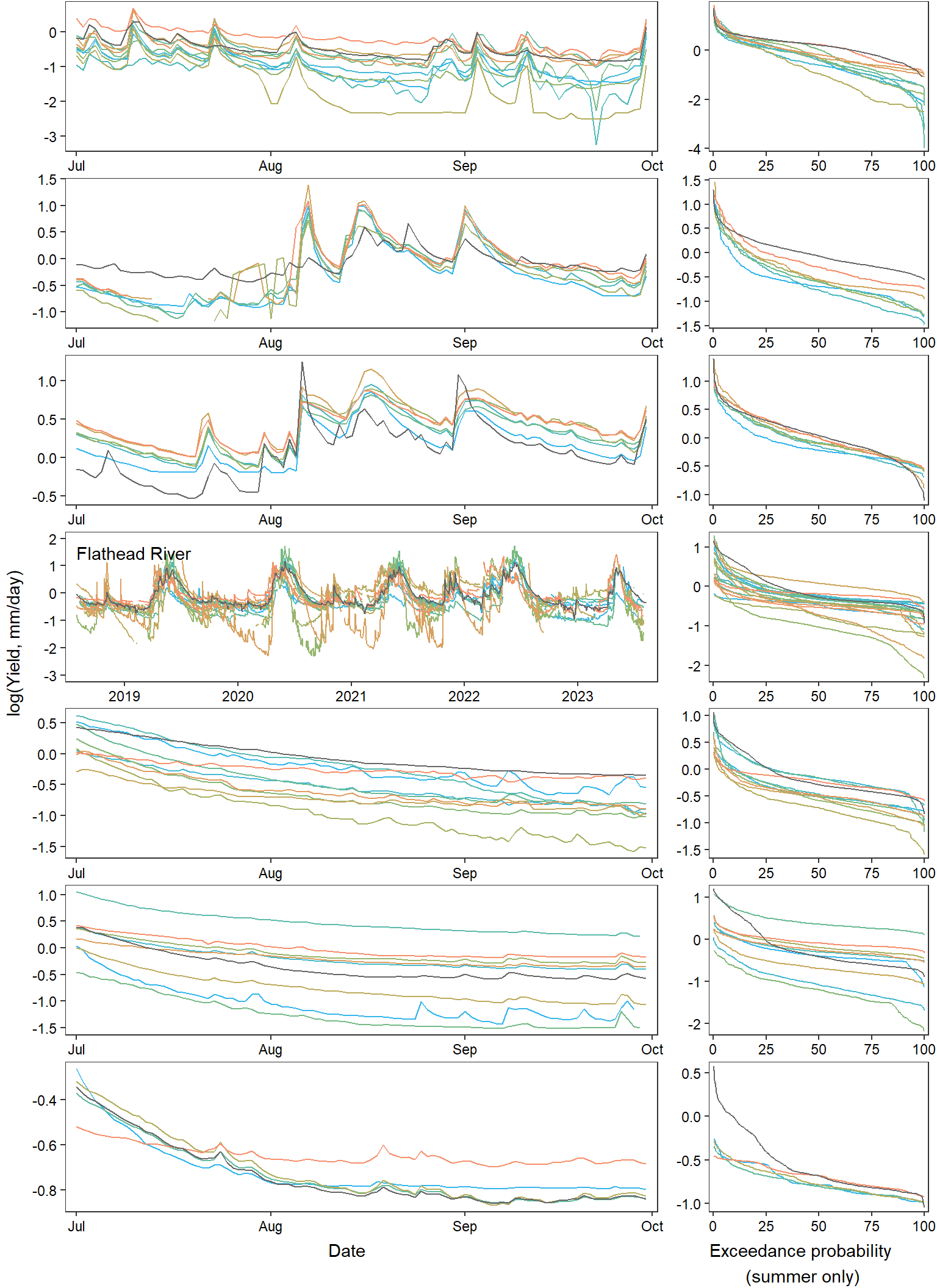
Write to file
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/EcoD_tsexed_new.jpg", width = 8, height = 11, units = "in", res = 1000)
p
dev.off()png
2 Use rose/radial plots to show differences in key flow metrics among sites within the West Brook. While these metrics/plots provide helpful summaries of flow regimes and variation among sites, extending to other regions/basins is not possible given the extent of seasonal/missing data.
Calculate flow metrics for each site:
Generate plot
Use CVPE (mean-variance Portfolio Effect, Anderson et al. 2013) to explore the effect site-level heterogeneity in flow regimes on network
Plotting function
# set up color palette
mycols <- c(brewer.pal(8, "Dark2"), "dodgerblue", "darkorchid")
# create plotting function
myplotfun <- function(subbas, lab, bigG) {
data_sub <- dat_clean %>% filter(subbasin == subbas)
par(mar = c(1.7,1.7,0.1,0.1), mgp = c(2.5,0.6,0))
plot(logYield ~ date, data_sub, type = "n", xlab = "", ylab = "")
#tempdat_little <- data_sub %>% filter(designation == "little")
tempdat_little <- fill_missing_dates(data_sub, dates = date, groups = site_name, pad_ends = FALSE)
tempdat_big <- dat_clean_big %>% filter(site_name == bigG, date >= min(tempdat_little$date), date <= max(tempdat_little$date))
#tempdat_big <- data_sub %>% filter(designation == "big")
tempdat_big <- fill_missing_dates(tempdat_big, dates = date, groups = site_name, pad_ends = FALSE)
mysites <- sort(unique(tempdat_little$site_name))
for (i in 1:length(mysites)) {
lines(logYield ~ date, tempdat_little %>% filter(site_name == mysites[i]), col = mycols[i], lwd = 0.7)
}
lines(logYield ~ date, tempdat_big, col = "white", lwd = 1.7)
lines(logYield ~ date, tempdat_big, col = "black", lwd = 1)
usr <- par("usr")
par(usr = c(0,1,0,1))
text(0.02, 0.9, labels = lab, cex = 1.2)
par(usr = usr)
}Generate plot
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/EcoD_timeseries.jpg", width = 8, height = 12, units = "in", res = 1000)
par(mfrow = c(10,1), oma = c(0.5,2,0.5,0.5))
# West Brook
myplotfun(subbas = "West Brook", lab = "(a)", bigG = "South River Conway NWIS")
# Paine
myplotfun(subbas = "Paine Run", lab = "(b)", bigG = "South River Harriston NWIS")
# Staunton River
myplotfun(subbas = "Staunton River", lab = "(c)", bigG = "Rapidan River NWIS")
# Big Creek
myplotfun(subbas = "Big Creek", lab = "(d)", bigG = "North Fork Flathead River NWIS")
# Coal Creek
myplotfun(subbas = "Coal Creek", lab = "(e)", bigG = "North Fork Flathead River NWIS")
# McGee Creek
myplotfun(subbas = "McGee Creek", lab = "(f)", bigG = "North Fork Flathead River NWIS")
# Snake River
myplotfun(subbas = "Snake River", lab = "(g)", bigG = "Pacific Creek at Moran NWIS")
# Shields River
myplotfun(subbas = "Shields River", lab = "(h)", bigG = "Yellowstone River Livingston NWIS")
# Duck Creek
myplotfun(subbas = "Duck Creek", lab = "(i)", bigG = "Yellowstone River Livingston NWIS")
# Donner Blitzen
myplotfun(subbas = "Donner Blitzen", lab = "(j)", bigG = "Donner Blitzen River nr Frenchglen NWIS")
# common axis label
mtext("log(Daily yield, mm)", outer = TRUE, side = 2, line = 0.5)
dev.off()Plotting functions
# create plotting function
myplotfun_ex <- function(subbas, lab) {
# filter to subbasin and get site designation
}
# CV of little g Yield by exceedance probability
myplotfun_cv <- function(subbas, lab) {
# filter to subbasin and get site designation
data_sub <- dat_clean %>% filter(subbasin == subbas)
sitesdesig <- data_sub %>% group_by(site_name) %>% summarize(designation = unique(designation)) %>% ungroup()
# calculate exceedance probability by site
exeeddat <- data_sub %>%
select(date, site_name, logYield) %>%
spread(key = site_name, value = logYield) %>%
drop_na() %>%
gather(key = site_name, value = logYield, 2:ncol(.)) %>%
group_by(site_name) %>%
arrange(desc(logYield), .by_group = TRUE) %>%
mutate(exceedance = 100/length(logYield)*1:length(logYield)) %>%
ungroup() %>%
left_join(sitesdesig)
exeeddat_little <- exeeddat %>% filter(designation == "little")
exeeddat_big <- exeeddat %>% filter(designation == "big")
# set up plot
par(mar = c(1.7,1.7,0.1,0.1), mgp = c(2.5,0.6,0))
mysites <- sort(unique(exeeddat_little$site_name))
tiblist <- list()
for (i in 1:length(mysites)) {
tt <- exeeddat %>% filter(site_name == mysites[i])
mylinint <- approx(x = tt$exceedance, y = tt$logYield, xout = seq(from = 0, to = 100, by = 1))
tiblist[[i]] <- tibble(site_name = mysites[i], exceedance = mylinint$x, logYield = mylinint$y)
}
cvtib <- do.call(bind_rows, tiblist) %>%
group_by(exceedance) %>%
summarize(sdf = sd(logYield)) %>%
ungroup()
plot(sdf ~ exceedance, cvtib, type = "l", col = "grey40", lwd = 2, ylim = c(0,0.8))
# panel label
usr <- par("usr")
par(usr = c(0,1,0,1))
text(0.92, 0.9, labels = lab, cex = 1.2)
par(usr = usr)
} Plot exceedance
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/EcoD_exceedance_JAS.jpg", width = 4, height = 8, units = "in", res = 1000)
par(mfrow = c(5,2), oma = c(2,2,0.5,0.5))
myplotfun_ex(subbas = "West Brook", lab = "(a)")
myplotfun_ex(subbas = "Paine Run", lab = "(b)")
myplotfun_ex(subbas = "Staunton River", lab = "(c)")
myplotfun_ex(subbas = "Big Creek", lab = "(d)")
myplotfun_ex(subbas = "Coal Creek", lab = "(e)")
myplotfun_ex(subbas = "McGee Creek", lab = "(f)")
myplotfun_ex(subbas = "Snake River", lab = "(g)")
myplotfun_ex(subbas = "Shields River", lab = "(h)")
myplotfun_ex(subbas = "Duck Creek", lab = "(i)")
myplotfun_ex(subbas = "Donner Blitzen", lab = "(j)")
mtext("log(Daily yield, mm)", outer = TRUE, side = 2, line = 0.5)
mtext("Exceedance probability", outer = TRUE, side = 1, line = 0.5)
dev.off()
#Plot CV by exceedance
jpeg("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/EcoD_exceedance_cv.jpg", width = 4, height = 8, units = "in", res = 1000)
par(mfrow = c(5,2), oma = c(2,2,0.5,0.5))
myplotfun_cv(subbas = "West Brook", lab = "(a)")
myplotfun_cv(subbas = "Paine Run", lab = "(b)")
myplotfun_cv(subbas = "Staunton River", lab = "(c)")
myplotfun_cv(subbas = "Big Creek", lab = "(d)")
myplotfun_cv(subbas = "Coal Creek", lab = "(e)")
myplotfun_cv(subbas = "McGee Creek", lab = "(f)")
myplotfun_cv(subbas = "Snake River", lab = "(g)")
myplotfun_cv(subbas = "Shields River", lab = "(h)")
myplotfun_cv(subbas = "Duck Creek", lab = "(i)")
myplotfun_cv(subbas = "Donner Blitzen", lab = "(j)")
mtext("SD of log(daily yield, mm) at little g's", outer = TRUE, side = 2, line = 0.5)
mtext("Exceedance probability", outer = TRUE, side = 1, line = 0.5)
dev.off()
#