Code
siteinfo <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Data/EcoDrought_SiteInformation.csv")
siteinfo_sp <- st_as_sf(siteinfo, coords = c("long", "lat"), crs = 4326)Purpose: Explore non-linear and hysteretic relationship between flow at reference vs. at headwater gages
Approach:
Notes:
Site information
siteinfo <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/Data/EcoDrought_SiteInformation.csv")
siteinfo_sp <- st_as_sf(siteinfo, coords = c("long", "lat"), crs = 4326)Little g’s
dat_clean <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/LittleG_data_clean.csv")Big G’s
dat_clean_big <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/BigG_data_clean.csv")Climate
climdf <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/Daymet_climate.csv")
climdf_summ <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/Daymet_climate_summary.csv")Water availability
wateravail <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/BigG_wateravailability_annual.csv")Find little g site/water years with at least 90% data completeness, join big and little g daily yield data
# find little g site/water years with at least 90% data availability
mysiteyrs <- dat_clean %>%
group_by(site_name, basin, subbasin, region, WaterYear) %>%
summarize(numdays = n(),
totalyield_site = sum(Yield_mm)) %>%
ungroup() %>%
filter(numdays >= 0.9*365) %>%
mutate(forhyst = 1) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield_z))
# join big G data to little g
# dat_clean_hyst <- dat_clean %>%
# left_join(mysiteyrs) %>%
# filter(forhyst == 1) %>%
# left_join(dat_clean_big %>% select(basin, date, Yield_mm, logYield) %>% rename(Yield_mm_big = Yield_mm, logYield_big = logYield)) %>%
# left_join(wateravail %>% select(basin, WaterYear, totalyield_z))
hystlist <- list()
for (i in 1:dim(mysiteyrs)[1]) {
tt <- dat_clean %>% filter(site_name == mysiteyrs$site_name[i], WaterYear == mysiteyrs$WaterYear[i])
tt <- fill_missing_dates(tt, dates = date, water_year_start = 10, pad_ends = TRUE)
tt <- tt %>%
mutate(site_name = mysiteyrs$site_name[i],
basin = mysiteyrs$basin[i],
subbasin = mysiteyrs$subbasin[i],
region = mysiteyrs$region[i]) %>%
select(site_name, basin, subbasin, region, date, Yield_mm, logYield)
tt <- add_date_variables(tt, dates = date, water_year_start = 10)
hystlist[[i]] <- tt %>%
left_join(dat_clean_big %>%
select(basin, date, Yield_mm, logYield) %>%
rename(Yield_mm_big = Yield_mm, logYield_big = logYield)) %>%
mutate(logYield_big = na.approx(logYield_big)) %>%
left_join(wateravail %>% select(basin, WaterYear, totalyield_z))
}
dat_clean_hyst <- do.call(rbind, hystlist)
# view data
mysiteyrs# A tibble: 87 × 9
site_name basin subbasin region WaterYear numdays totalyield_site forhyst
<chr> <chr> <chr> <chr> <dbl> <int> <dbl> <dbl>
1 Avery Brook West… West Br… Mass 2021 365 872. 1
2 Avery Brook West… West Br… Mass 2022 365 602. 1
3 Avery Brook West… West Br… Mass 2023 365 1147. 1
4 Avery Brook West… West Br… Mass 2024 345 993. 1
5 Big Creek NW… Flat… Big Cre… Flat 2019 329 566. 1
6 Big Creek NW… Flat… Big Cre… Flat 2020 347 719. 1
7 Big Creek NW… Flat… Big Cre… Flat 2021 352 554. 1
8 Big Creek NW… Flat… Big Cre… Flat 2022 340 842. 1
9 Buck Creek Shie… Shields… Shiel… 2022 329 174. 1
10 CycloneCreek… Flat… Coal Cr… Flat 2019 354 374. 1
# ℹ 77 more rows
# ℹ 1 more variable: totalyield_z <dbl>dat_clean_hyst# A tibble: 31,778 × 15
site_name basin subbasin region date Yield_mm logYield CalendarYear
<chr> <chr> <chr> <chr> <date> <dbl> <dbl> <dbl>
1 Avery Brook West B… West Br… Mass 2020-10-01 0.595 -0.226 2020
2 Avery Brook West B… West Br… Mass 2020-10-02 0.341 -0.467 2020
3 Avery Brook West B… West Br… Mass 2020-10-03 0.288 -0.541 2020
4 Avery Brook West B… West Br… Mass 2020-10-04 0.218 -0.662 2020
5 Avery Brook West B… West Br… Mass 2020-10-05 0.204 -0.689 2020
6 Avery Brook West B… West Br… Mass 2020-10-06 0.209 -0.679 2020
7 Avery Brook West B… West Br… Mass 2020-10-07 0.239 -0.621 2020
8 Avery Brook West B… West Br… Mass 2020-10-08 0.262 -0.581 2020
9 Avery Brook West B… West Br… Mass 2020-10-09 0.221 -0.657 2020
10 Avery Brook West B… West Br… Mass 2020-10-10 0.245 -0.611 2020
# ℹ 31,768 more rows
# ℹ 7 more variables: Month <dbl>, MonthName <fct>, WaterYear <dbl>,
# DayofYear <dbl>, Yield_mm_big <dbl>, logYield_big <dbl>, totalyield_z <dbl>Create plotting function
hystplotfun <- function(mysite, wy, months = c(1:12)) {
(dat_clean_hyst %>%
filter(site_name == mysite, WaterYear == wy, Month %in% months) %>%
ggplot(aes(x = logYield_big, y = logYield, color = date)) +
geom_segment(aes(xend = c(tail(logYield_big, n = -1), NA), yend = c(tail(logYield, n = -1), NA)), arrow = arrow(length = unit(0.2, "cm")), color = "black") +
geom_point() +
geom_abline(intercept = 0, slope = 1, linetype = "dashed") +
scale_color_gradientn(colors = cet_pal(250, name = "c2"), trans = "date") +
xlim(-1,1.5) + ylim(-1.5,1.8) +
#scale_color_scico(palette = "romaO") +
xlab("log(Yield, mm) at reference gage") + ylab("log(Yield, mm) at headwater gage") +
# geom_text(data = annotations, aes(x = xpos, y = ypos, label = annotateText), hjust = "inward", vjust = "inward") +
annotate(geom = "text", x = -Inf, y = Inf, label = paste(mysite, ", WY ", wy, sep = ""), hjust = -0.1, vjust = 1.5) +
theme_bw() +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_rect(fill = "grey85"),
axis.title = element_blank()) +
theme(plot.margin = margin(0.1,0.1,0,0, "cm")) )
}
# change to linear color palette
hystplotfunlin <- function(mysite, wy, months = c(1:12)) {
(dat_clean_hyst %>%
filter(site_name == mysite, WaterYear == wy, Month %in% months) %>%
ggplot(aes(x = logYield_big, y = logYield, color = date)) +
geom_segment(aes(xend = c(tail(logYield_big, n = -1), NA), yend = c(tail(logYield, n = -1), NA)), arrow = arrow(length = unit(0.2, "cm")), color = "black") +
geom_point() +
geom_abline(intercept = 0, slope = 1, linetype = "dashed") +
scale_color_gradientn(colors = cet_pal(90, name = "r1"), trans = "date") +
xlim(-1,1.5) + ylim(-1.5,1.8) +
#scale_color_scico(palette = "romaO") +
xlab("log(Yield, mm) at reference gage") + ylab("log(Yield, mm) at headwater gage") +
# geom_text(data = annotations, aes(x = xpos, y = ypos, label = annotateText), hjust = "inward", vjust = "inward") +
annotate(geom = "text", x = -Inf, y = Inf, label = paste(mysite, ", WY ", wy, sep = ""), hjust = -0.1, vjust = 1.5) +
theme_bw() +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_rect(fill = "grey85"),
axis.title = element_blank()) +
theme(plot.margin = margin(0.1,0.1,0,0, "cm")) )
}Plot example sites and years. Generally, in rain-dominated basins the East (top two rows), we see relatively little hysteresis/non-stationarity in the relationship between streamflow in headwaters and at reference gages. In contrast, in snowmelt-dominated basins of the Rocky Mountains, we see much stronger hysteresis/non-stationarity in the relationship between streamflow in headwaters and at reference gages, but this varies considerably among locations.
ggarrange(hystplotfun(mysite = "West Brook NWIS", wy = 2021) + theme(legend.position = "none"),
hystplotfun(mysite = "Jimmy Brook", wy = 2021) + theme(legend.position = "none"),
hystplotfun(mysite = "Sanderson Brook", wy = 2021) + theme(legend.position = "none"),
hystplotfun(mysite = "Avery Brook", wy = 2021) + theme(legend.position = "none"),
hystplotfun(mysite = "Staunton River 10", wy = 2021) + theme(legend.position = "none"),
hystplotfun(mysite = "Staunton River 06", wy = 2021) + theme(legend.position = "none"),
hystplotfun(mysite = "Staunton River 03", wy = 2021) + theme(legend.position = "none"),
hystplotfun(mysite = "Staunton River 02", wy = 2021) + theme(legend.position = "none"),
hystplotfun(mysite = "Big Creek NWIS", wy = 2020) + theme(legend.position = "none"),
hystplotfun(mysite = "WernerCreek", wy = 2020) + theme(legend.position = "none"),
hystplotfun(mysite = "CycloneCreekUpper", wy = 2020) + theme(legend.position = "none"),
hystplotfun(mysite = "McGeeCreekTrib", wy = 2020) + theme(legend.position = "none"),
hystplotfun(mysite = "Buck Creek", wy = 2022) + theme(legend.position = "none"),
hystplotfun(mysite = "Dugout Creek", wy = 2022) + theme(legend.position = "none"),
hystplotfun(mysite = "EF Duck Creek be HF", wy = 2022) + theme(legend.position = "none"),
get_legend(hystplotfun(mysite = "EF Duck Creek be HF", wy = 2022)),
nrow = 4, ncol = 4) %>%
annotate_figure(left = text_grob("log(Yield, mm) at headwater gage", rot = 90),
bottom = text_grob("log(Yield, mm) at reference gage"))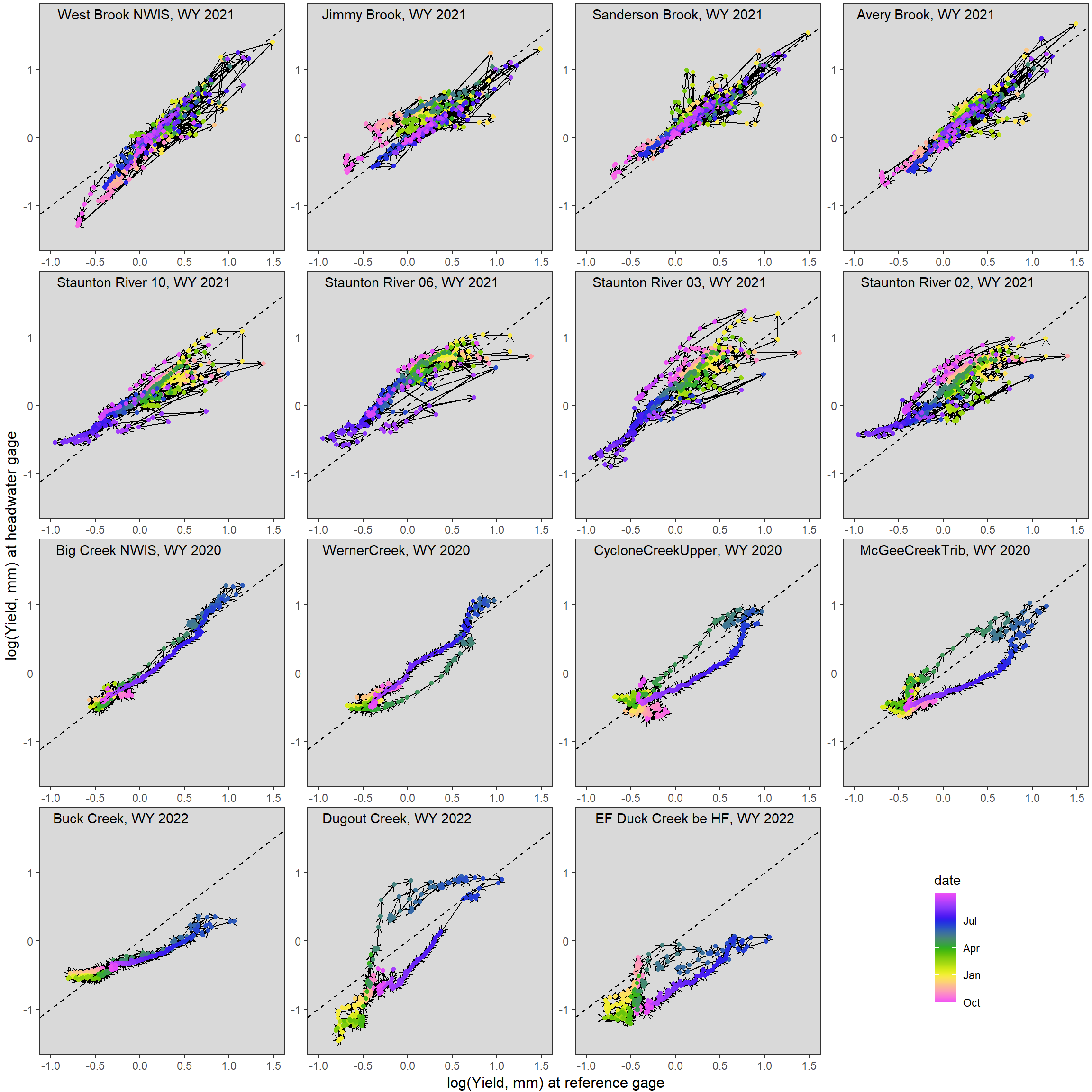
What if we just consider the summer (low flow) period, July - September? In some ways, we see an opposite pattern relative to that describe above. In that we see greater hysteretic behavior in rain-dominated basins (driven by frequent summer storms and lagged runoff response) and less hysteretic behavior in snow-dominated basins, as all streams are in recession from the spring snowmelt peak. However, in general, we see greater divergence from a 1:1 relationship in snowmelt vs rain-dominated basins.
ggarrange(hystplotfunlin(mysite = "West Brook NWIS", wy = 2021, months = c(7:9)) + theme(legend.position = "none"),
hystplotfunlin(mysite = "Jimmy Brook", wy = 2021, months = c(7:9)) + theme(legend.position = "none"),
hystplotfunlin(mysite = "Sanderson Brook", wy = 2021, months = c(7:9)) + theme(legend.position = "none"),
hystplotfunlin(mysite = "Avery Brook", wy = 2021, months = c(7:9)) + theme(legend.position = "none"),
hystplotfunlin(mysite = "Staunton River 10", wy = 2021, months = c(7:9)) + theme(legend.position = "none"),
hystplotfunlin(mysite = "Staunton River 06", wy = 2021, months = c(7:9)) + theme(legend.position = "none"),
hystplotfunlin(mysite = "Staunton River 03", wy = 2021, months = c(7:9)) + theme(legend.position = "none"),
hystplotfunlin(mysite = "Staunton River 02", wy = 2021, months = c(7:9)) + theme(legend.position = "none"),
hystplotfunlin(mysite = "Big Creek NWIS", wy = 2020, months = c(7:9)) + theme(legend.position = "none"),
hystplotfunlin(mysite = "WernerCreek", wy = 2020, months = c(7:9)) + theme(legend.position = "none"),
hystplotfunlin(mysite = "CycloneCreekUpper", wy = 2020, months = c(7:9)) + theme(legend.position = "none"),
hystplotfunlin(mysite = "McGeeCreekTrib", wy = 2020, months = c(7:9)) + theme(legend.position = "none"),
hystplotfunlin(mysite = "Buck Creek", wy = 2022, months = c(7:9)) + theme(legend.position = "none"),
hystplotfunlin(mysite = "Dugout Creek", wy = 2022, months = c(7:9)) + theme(legend.position = "none"),
hystplotfunlin(mysite = "EF Duck Creek be HF", wy = 2022, months = c(7:9)) + theme(legend.position = "none"),
get_legend(hystplotfunlin(mysite = "EF Duck Creek be HF", wy = 2022, months = c(7:9))),
nrow = 4, ncol = 4) %>%
annotate_figure(left = text_grob("log(Yield, mm) at headwater gage", rot = 90),
bottom = text_grob("log(Yield, mm) at reference gage"))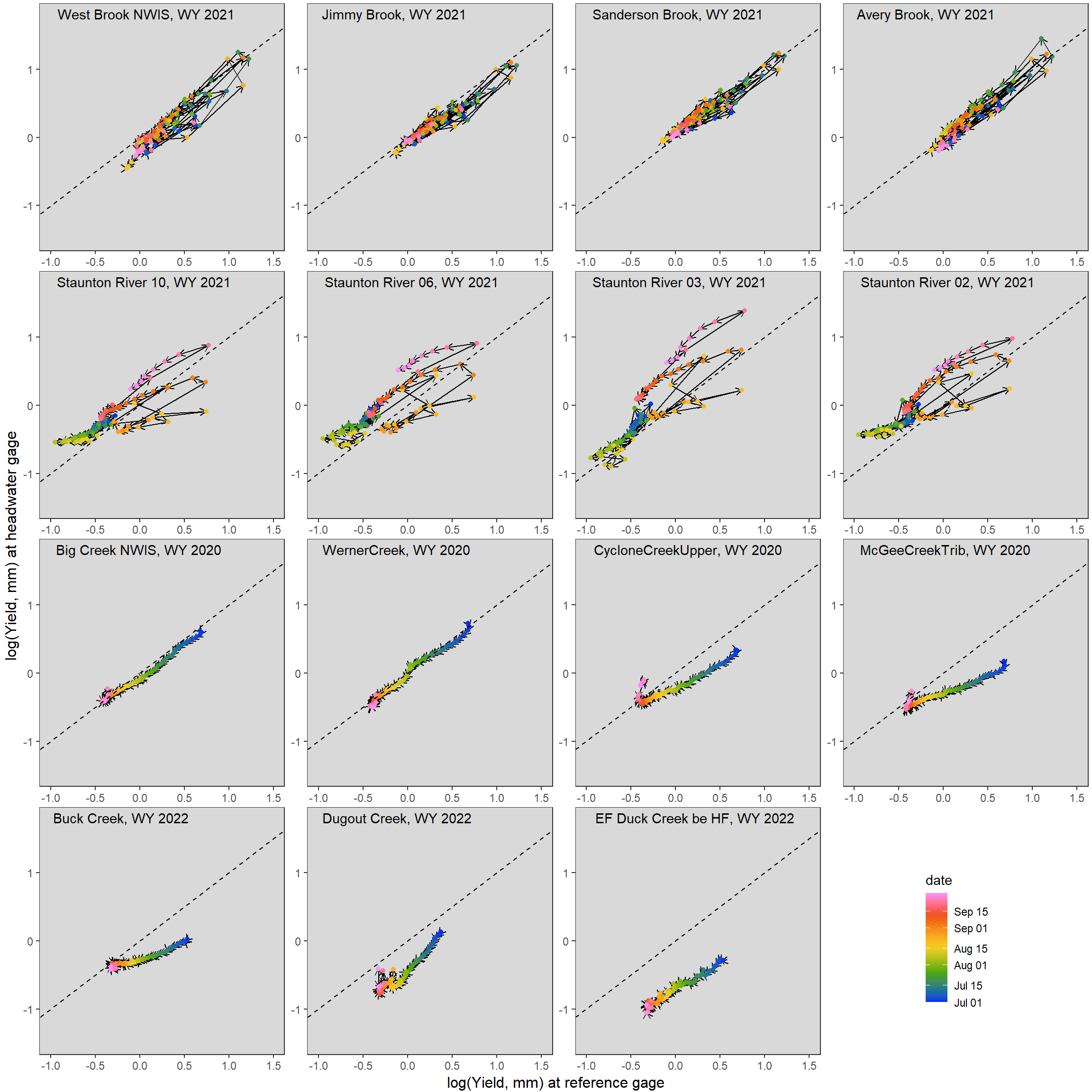
ggplotly(hystplotfun(mysite = "West Brook NWIS", wy = 2021))ggplotly(hystplotfun(mysite = "Jimmy Brook", wy = 2021))ggplotly(hystplotfun(mysite = "Sanderson Brook", wy = 2021))ggplotly(hystplotfun(mysite = "Avery Brook", wy = 2021))ggplotly(hystplotfun(mysite = "Staunton River 10", wy = 2021))ggplotly(hystplotfun(mysite = "Staunton River 06", wy = 2021))ggplotly(hystplotfun(mysite = "Staunton River 03", wy = 2021))ggplotly(hystplotfun(mysite = "Staunton River 02", wy = 2021))ggplotly(hystplotfun(mysite = "Big Creek NWIS", wy = 2020))ggplotly(hystplotfun(mysite = "WernerCreek", wy = 2020))ggplotly(hystplotfun(mysite = "CycloneCreekUpper", wy = 2020))ggplotly(hystplotfun(mysite = "McGeeCreekTrib", wy = 2020))ggplotly(hystplotfun(mysite = "Buck Creek", wy = 2022))ggplotly(hystplotfun(mysite = "Dugout Creek", wy = 2022))ggplotly(hystplotfun(mysite = "EF Duck Creek be HF", wy = 2022))Calculate index of hystereis for each event and characterize size-years according the frequency of events and magnitude of hysteretic effects.
Conduct baseflow extraction, event delineation, and calculate event-specific hysteresis index using dynamic time warping (Xue et al. 2024).
# set baseflow extraction and event delineation parameters
alp <- 0.95
numpass <- 3
thresh <- 0.75
# empty list to story site-year data
hysteresis_list <- list()
for (j in 1:nrow(mysiteyrs)) {
# filter data by site name and water year
dd <- dat_clean_hyst %>% filter(site_name == mysiteyrs$site_name[j], WaterYear == mysiteyrs$WaterYear[j])
# baseflow extraction on big G yield
dd <- dd %>%
filter(!is.na(Yield_mm_big)) %>%
mutate(bf = baseflowB(Yield_mm_big, alpha = alp, passes = numpass)$bf,
bfi = baseflowB(Yield_mm_big, alpha = alp, passes = numpass)$bfi) %>%
ungroup()
# delineate events
events <- eventBaseflow(dd$Yield_mm_big, BFI_Th = thresh, bfi = dd$bfi)
events <- events %>% mutate(len = end - srt + 1)
#print("Done - baseflow extraction and event delineation")
## Tidy events
# define positions of non-events
# srt <- c(1)
# end <- c(events$srt[1]-1)
# for (i in 2:(dim(events)[1])) {
# srt[i] <- events$end[i-1]+1
# end[i] <- events$srt[i]-1
# }
# nonevents <- data.frame(tibble(srt, end) %>%
# mutate(len = end - srt) %>%
# filter(len >= 0) %>% select(-len) %>%
# add_row(srt = events$end[dim(events)[1]]+1, end = dim(dd)[1])
# )
# create vectors of binary event/non-event and event IDs
isevent_vec <- rep(2, times = dim(dd)[1])
eventid_vec <- rep(NA, times = dim(dd)[1])
for (i in 1:dim(events)[1]) {
isevent_vec[c(events[i,1]:events[i,2])] <- 1
eventid_vec[c(events[i,1]:events[i,2])] <- i
}
# # create vector of non-event IDs
# noneventid_vec <- rep(NA, times = dim(dd)[1])
# for (i in 1:dim(nonevents)[1]) { noneventid_vec[c(nonevents[i,1]:nonevents[i,2])] <- i }
#
# # create vector of "agnostic events": combined hydro events and non-events
# agnevents <- rbind(events %>% select(srt, end) %>% mutate(event = 1), nonevents %>% mutate(event = 0)) %>% arrange((srt))
# agneventid_vec <- c()
# for (i in 1:dim(agnevents)[1]){ agneventid_vec[c(agnevents[i,1]:agnevents[i,2])] <- i }
# add event/non-event vectors to Big G data
dd <- dd %>%
mutate(isevent = isevent_vec,
eventid = eventid_vec,
#noneventid = noneventid_vec,
#agneventid = agneventid_vec,
big_event_yield = ifelse(isevent_vec == 1, Yield_mm_big, NA),
big_nonevent_yield = ifelse(isevent_vec == 2, Yield_mm_big, NA),
big_event_quick = big_event_yield - bf) %>%
rename(big_bf = bf, big_bfi = bfi)
#print("Done - tidy events")
# calculate hysteresis index for each event using dynamic time warping
event_list <- list()
for (i in 1:max(dd$eventid, na.rm = TRUE)) {
# filter by each event and normalized big and little Yield
ddd <- dd %>% filter(eventid == i, !is.na(logYield), !is.na(logYield_big))
dddd <- dd %>% filter(date %in% c(min(ddd$date)-1, max(ddd$date)+1))
ddd <- ddd %>%
bind_rows(dddd) %>%
arrange(date) %>%
mutate(weight = (logYield_big - min(logYield_big)) / (max(logYield_big) - min(logYield_big)),
yield_little_norm = (logYield - min(logYield)) / (max(logYield) - min(logYield)),
yield_big_norm = (logYield_big - min(logYield_big)) / (max(logYield_big) - min(logYield_big)))
# ddd %>%
# ggplot(aes(x = yield_big_norm, y = yield_little_norm, color = date)) +
# geom_segment(aes(xend = c(tail(yield_big_norm, n = -1), NA), yend = c(tail(yield_little_norm, n = -1), NA)),
# arrow = arrow(length = unit(0.2, "cm")), color = "black") +
# geom_point() +
# geom_abline(intercept = 0, slope = 1, linetype = "dashed") +
# scale_color_gradientn(colors = cet_pal(250, name = "r1"), trans = "date") +
# theme_bw() +
# theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
# align data using dynamic time warping, allow for error handling (e.g., skips if either big or little g is constant during event, b/c normalization returns NaN)
skip_to_next <- FALSE
tryCatch(align <- dtw(x = unlist(ddd %>% select(yield_big_norm)), y = unlist(ddd %>% select(yield_little_norm)), step = asymmetric, keep = TRUE), error = function(e) { skip_to_next <<- TRUE })
if(skip_to_next) { next }
#align <- dtw(x = unlist(ddd %>% select(yield_big_norm)), y = unlist(ddd %>% select(yield_little_norm)), step = asymmetric, keep = TRUE)
# plot(align, type = "threeway")
# plot(align, type = "twoway", offset = -1)
# find time-warped distance, S_i in Xue et al. (2024)
ddd <- ddd %>% mutate(distance = align$index1 - align$index2)
# ddd %>%
# ggplot(aes(x = date, y = distance*weight)) +
# geom_bar(stat = "identity") +
# theme_bw()
# summarize event
event_list[[i]] <- ddd %>%
group_by(site_name, basin, subbasin, region, WaterYear) %>%
summarize(eventdays = n(),
eventid = i,
mindate = min(date),
maxdate = max(date),
totalyieldevent_little = sum(Yield_mm),
totalyieldevent_big = sum(Yield_mm_big),
hysteresis = sum(ddd$weight * ddd$distance) / sum(ddd$weight) # hysteresis index as in Xue et al (2024)
) %>%
ungroup()
}
#print("Done - dynamic time warping")
hysteresis_list[[j]] <- do.call(bind_rows, event_list)
print(j)
}
# bind to tibble
hysteresis <- do.call(bind_rows, hysteresis_list) %>%
left_join(mysiteyrs %>% select(site_name, basin, subbasin, region, WaterYear, numdays, totalyield_site)) %>%
mutate(propyield = totalyieldevent_little / totalyield_site)
# write to file
write_csv(hysteresis, "C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/hysteresis.csv")Load hysteresis file
hysteresis <- read_csv("C:/Users/jbaldock/OneDrive - DOI/Documents/USGS/EcoDrought/EcoDrought Working/EcoDrought-Analysis/Qualitative/hysteresis.csv")
# hysteresis <- hysteresis %>%
# group_by(subbasin, WaterYear) %>%
# mutate(eventid = as.numeric(as.factor(mindate))) %>%
# ungroup()Distribution of hystersis index by subbasin
hysteresis %>%
ggplot(aes(x = subbasin, y = hysteresis, fill = subbasin)) +
geom_boxplot(outlier.shape = NA) +
geom_jitter(height = 0, width = 0.25, alpha = 0.2) +
xlab("Sub-basin") + ylab("Xue hysteresis index") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
axis.text.x = element_text(angle = 45, vjust = 0.5, hjust = 0.4))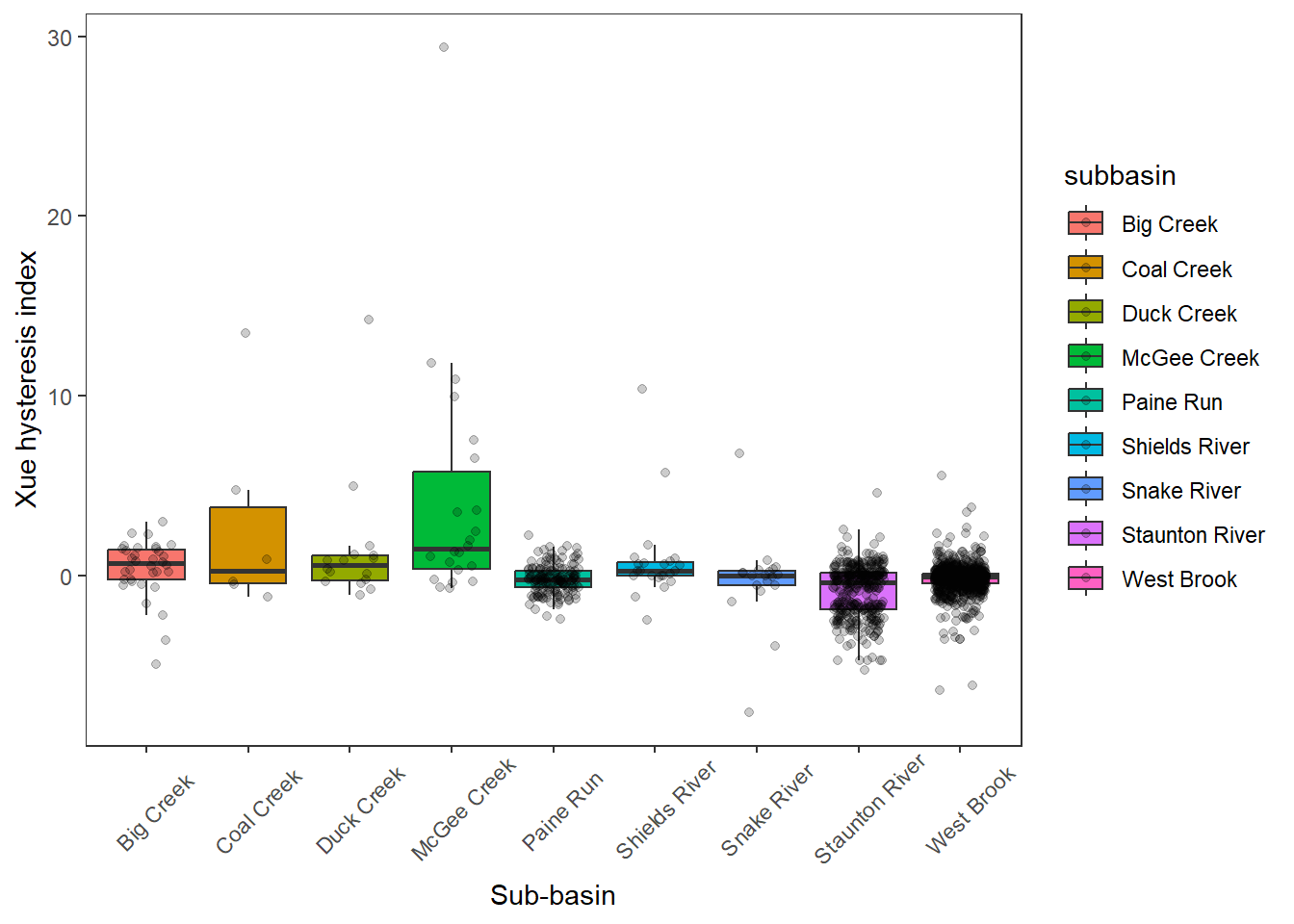
Magnitude of hysteresis by length of event
hysteresis %>%
ggplot(aes(x = eventdays, y = abs(hysteresis), color = subbasin)) +
geom_point() +
xlab("Event length (days)") + ylab("Xue hysteresis index (absolute value)") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())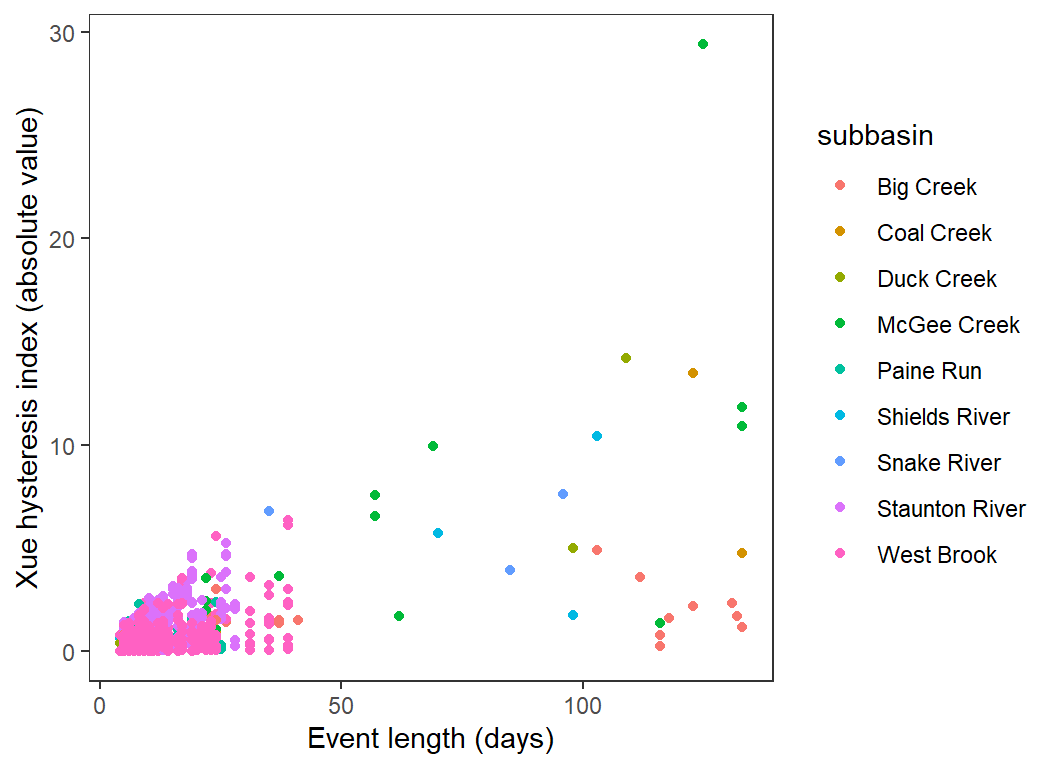
Magnitude of hysteresis by length of event, by subbasin and site
hysteresis %>%
#filter(eventdays >= 6) %>%
ggplot(aes(x = eventdays, y = abs(hysteresis), color = site_name, group = site_name)) +
geom_point() +
geom_smooth(method = "lm") +
facet_wrap(~subbasin, scales = "free") +
xlab("Event length (days)") + ylab("Xue hysteresis index (absolute value)") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none")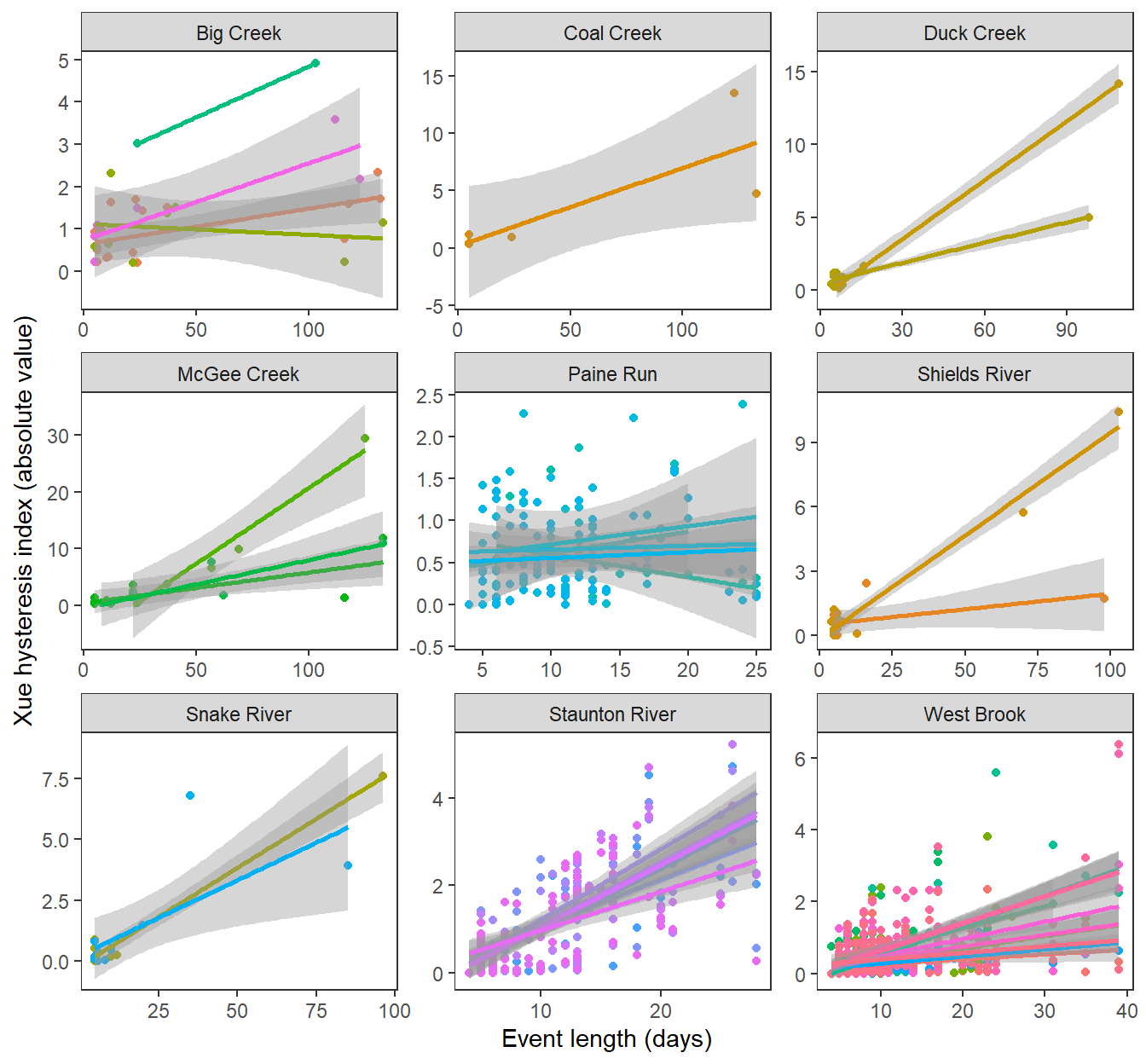
Magnitude of hysteresis by proportion of annual yield in event
hysteresis %>%
ggplot(aes(x = propyield, y = abs(hysteresis), color = subbasin)) +
geom_point() +
xlab("Proportion of annual yield in event") + ylab("Xue hysteresis index (absolute value)") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())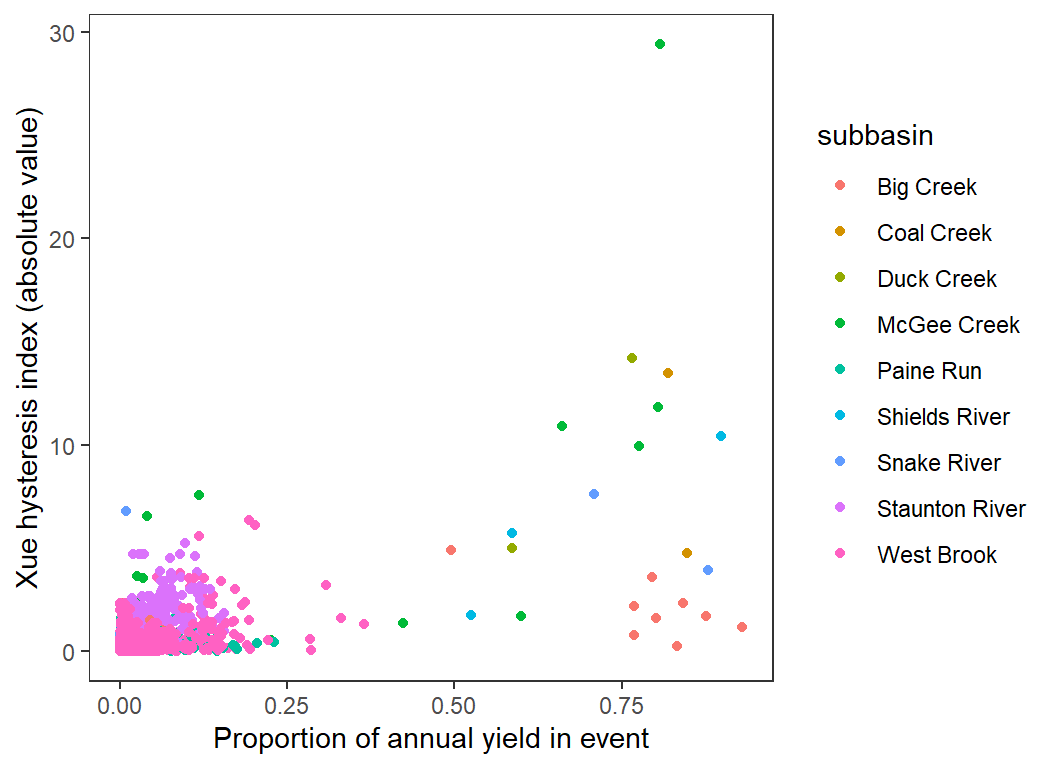
Magnitude of hysteresis by proportion of annual yield in event, by subbasin and site
hysteresis %>%
ggplot(aes(x = propyield, y = abs(hysteresis), color = site_name, group = site_name)) +
geom_point() +
geom_smooth(method = "lm") +
facet_wrap(~subbasin, scales = "free") +
xlab("Proportion of annual yield in event") + ylab("Xue hysteresis index (absolute value)") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none")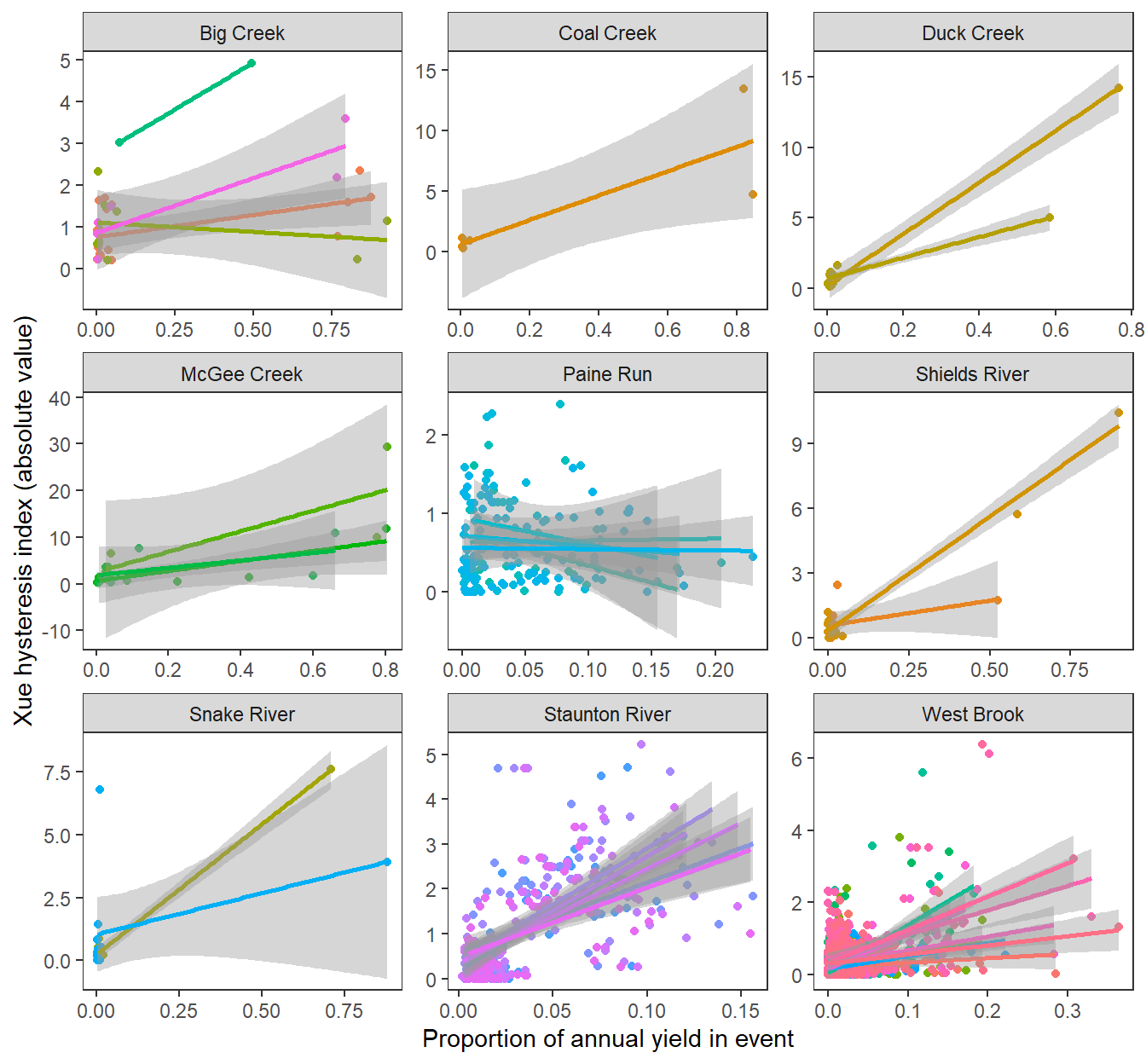
Characterize site-years along axes of mean annual event duration/magnitude and magnitude of hysteres.
Summarize hysteresis by site and year
temp <- hysteresis %>%
group_by(subbasin, site_name, WaterYear) %>%
summarize(freq = n(),
meanpropyield = mean(propyield),
meanhyst = mean(abs(hysteresis))) %>%
mutate(logmeanhyst = log(meanhyst)) %>%
ungroup()
temp# A tibble: 87 × 7
subbasin site_name WaterYear freq meanpropyield meanhyst logmeanhyst
<chr> <chr> <dbl> <int> <dbl> <dbl> <dbl>
1 Big Creek Big Creek NWIS 2019 2 0.435 2.02 0.703
2 Big Creek Big Creek NWIS 2020 3 0.294 0.954 -0.0473
3 Big Creek Big Creek NWIS 2021 6 0.143 0.878 -0.130
4 Big Creek Big Creek NWIS 2022 7 0.134 0.746 -0.294
5 Big Creek Hallowat Creek… 2020 2 0.465 0.868 -0.141
6 Big Creek Hallowat Creek… 2022 6 0.162 1.05 0.0452
7 Big Creek NicolaCreek 2019 2 0.284 3.96 1.38
8 Big Creek WernerCreek 2019 2 0.407 1.84 0.608
9 Big Creek WernerCreek 2020 4 0.202 1.43 0.358
10 Coal Creek CycloneCreekUp… 2019 2 0.436 2.84 1.04
# ℹ 77 more rowsCharacterize sites/subbasins along axes of event frequency and magnitude (mean annual proportion of flow within event).
hull <- temp %>% group_by(subbasin) %>% slice(chull(freq, meanpropyield))
temp %>%
ggplot(aes(x = freq, y = meanpropyield)) +
#geom_polygon(data = hull, aes(fill = subbasin), alpha = 0.5) +
#geom_smooth(color = "black", method = "lm") +
geom_point(aes(color = subbasin)) +
xlab("Events per year") + ylab("Proportion of annual yield in event (mean over events)") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())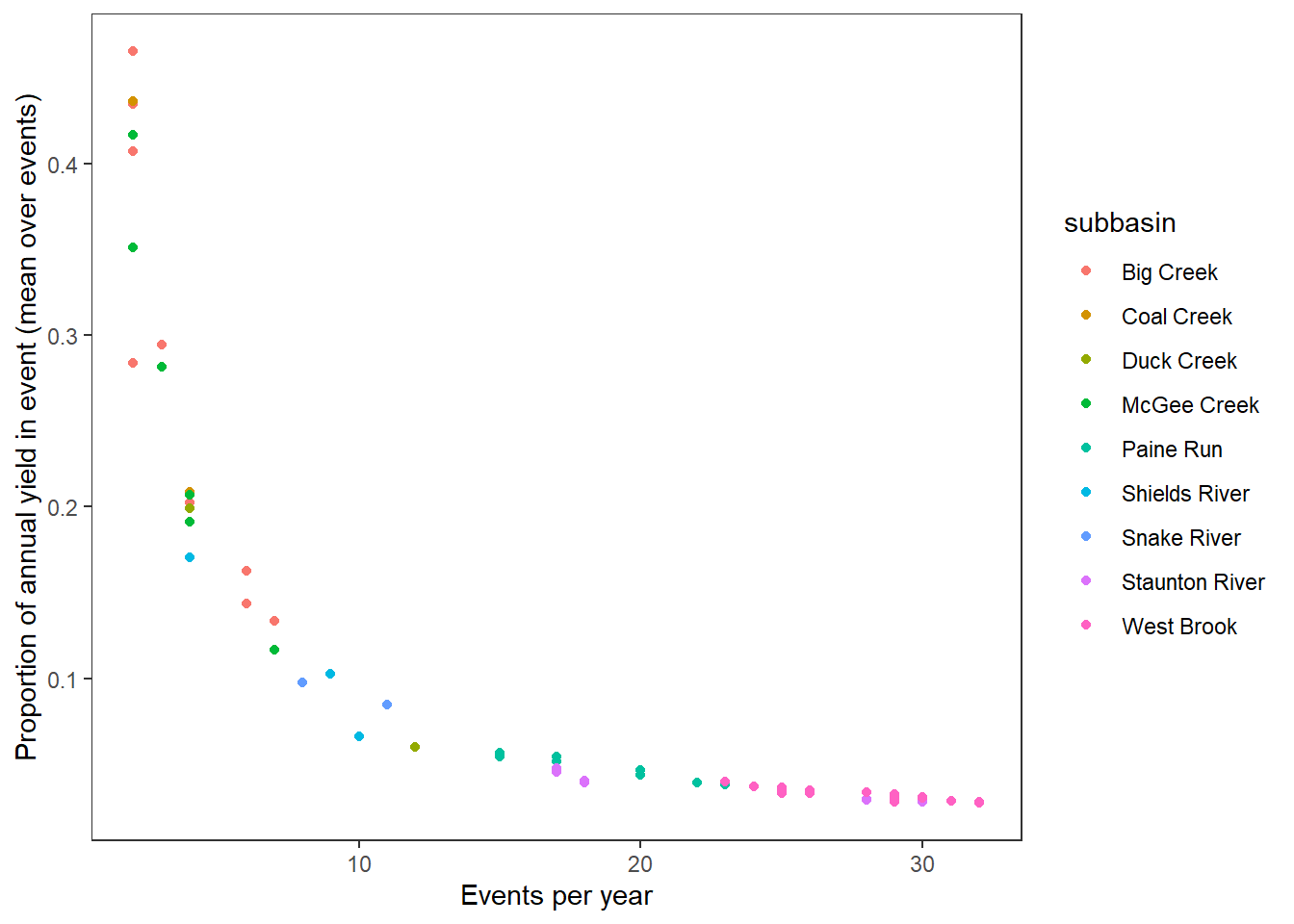
Plot the mean annual magnitude (absolute value) of event-specific hysteresis by event frequency (events per year), with LOESS smoothing.
p1 <- temp %>%
ggplot(aes(x = freq, y = (meanhyst))) +
geom_smooth(color = "black") +
#geom_smooth(color = "black", method = "glm", formula = y~x, method.args = list(family = gaussian(link = 'log'))) +
geom_point(aes(color = subbasin)) +
xlab("Events per year") + ylab("Mean annual magnitude of hysteresis") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
p2 <- temp %>%
ggplot(aes(x = meanpropyield, y = (meanhyst))) +
geom_smooth(color = "black") +
#geom_smooth(color = "black", method = "glm", formula = y~x, method.args = list(family = gaussian(link = 'log'))) +
geom_point(aes(color = subbasin)) +
xlab("Proportion of annual yield in event") + ylab("Mean annual magnitude of hysteresis") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
ggarrange(p1, p2, nrow = 1, common.legend = TRUE, legend = "right")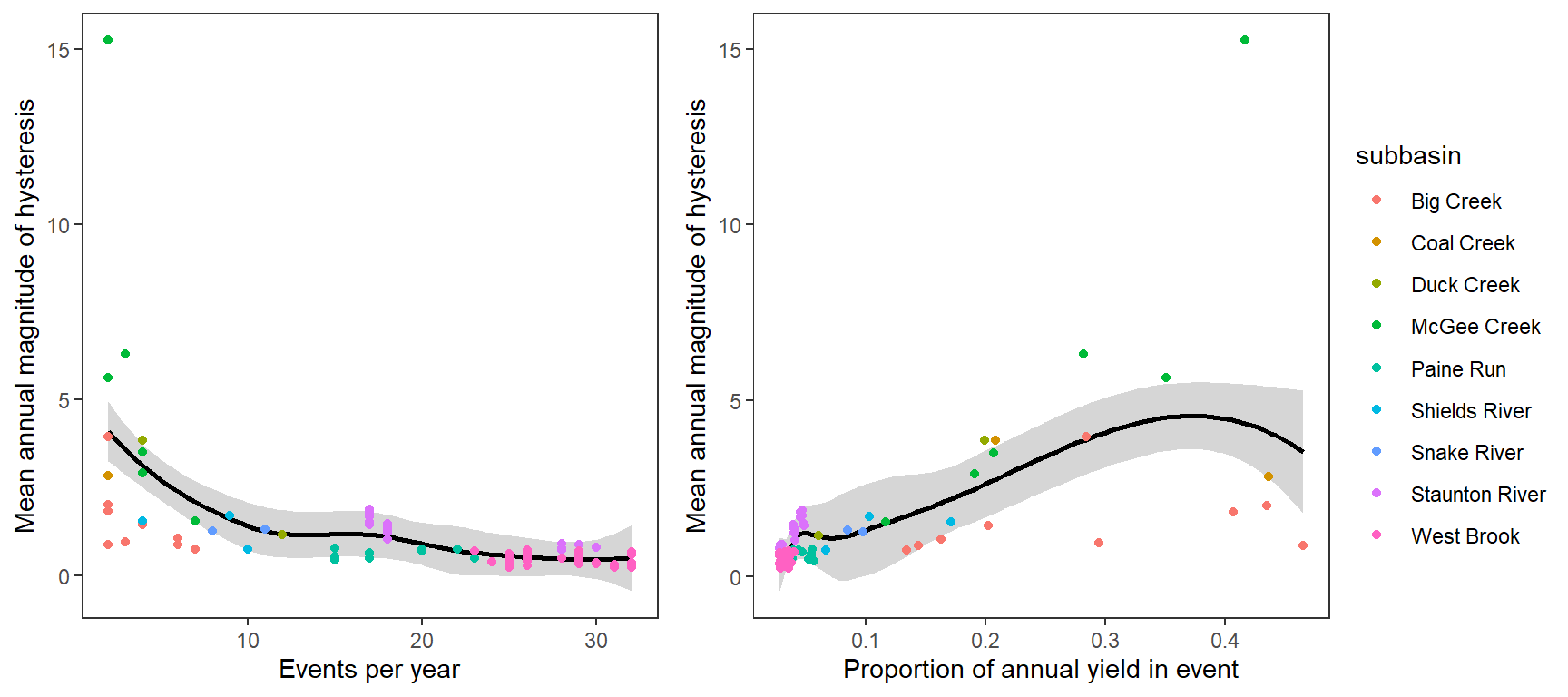
Same as above but plot on log scale (magnitude):
p1 <- temp %>%
ggplot(aes(x = freq, y = logmeanhyst)) +
geom_smooth(color = "black", method = "lm") +
geom_point(aes(color = subbasin)) +
xlab("Events per year") + ylab("(log) Mean annual magnitude of hysteresis") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
p2 <- temp %>%
ggplot(aes(x = meanpropyield, y = logmeanhyst)) +
geom_smooth(color = "black", method = "lm") +
geom_point(aes(color = subbasin)) +
xlab("Proportion of annual yield in event") + ylab("(log) Mean annual magnitude of hysteresis") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
ggarrange(p1, p2, nrow = 1, common.legend = TRUE, legend = "right")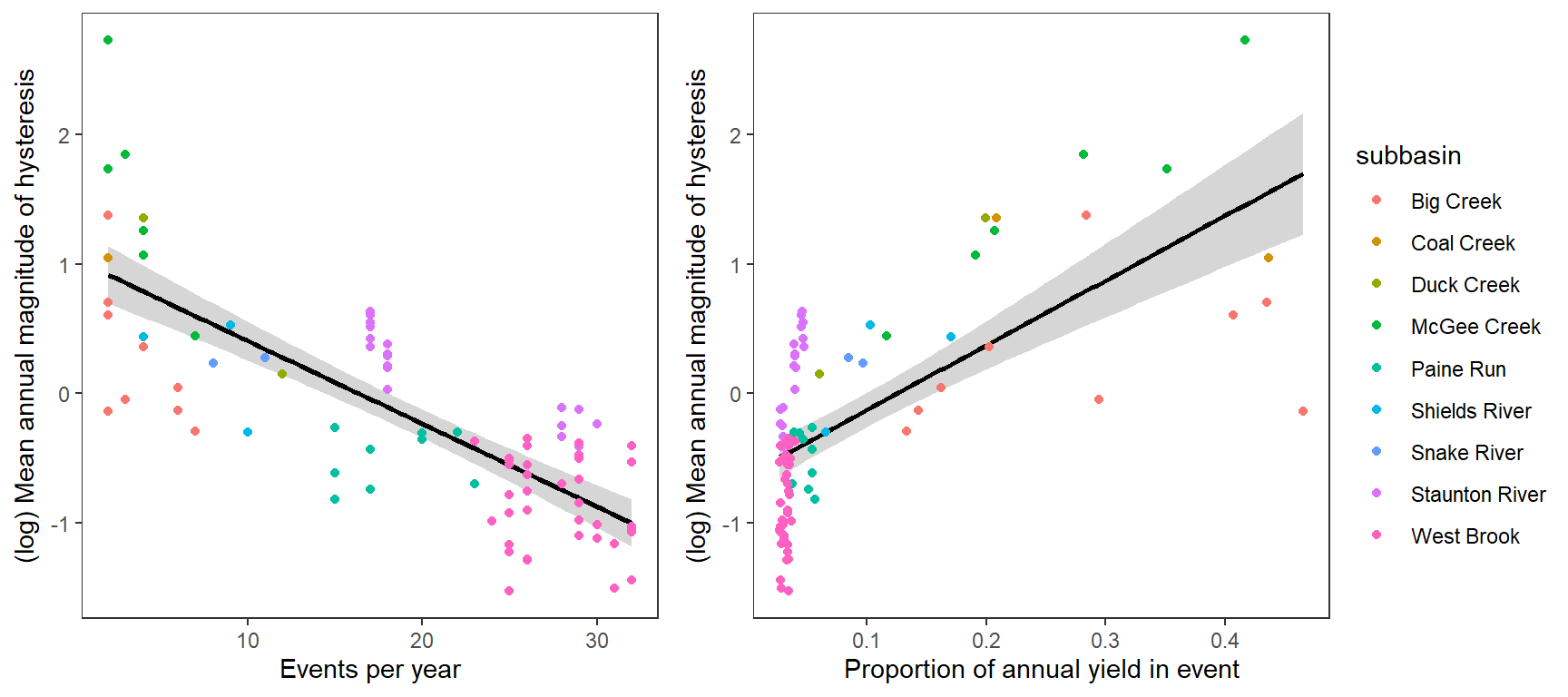
Replace regression line with convex hulls
hull <- temp %>% group_by(subbasin) %>% slice(chull(freq, logmeanhyst))
p1 <- temp %>%
ggplot(aes(x = freq, y = logmeanhyst)) +
geom_polygon(data = hull, aes(fill = subbasin), alpha = 0.4) +
geom_point(aes(color = subbasin)) +
xlab("Events per year") + ylab("(log) Mean annual magnitude of hysteresis") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
hull <- temp %>% group_by(subbasin) %>% slice(chull(meanpropyield, logmeanhyst))
p2 <- temp %>%
ggplot(aes(x = meanpropyield, y = logmeanhyst)) +
geom_polygon(data = hull, aes(fill = subbasin), alpha = 0.4) +
geom_point(aes(color = subbasin)) +
xlab("Proportion of annual yield in event") + ylab("(log) Mean annual magnitude of hysteresis") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
ggarrange(p1, p2, nrow = 1, common.legend = TRUE, legend = "right")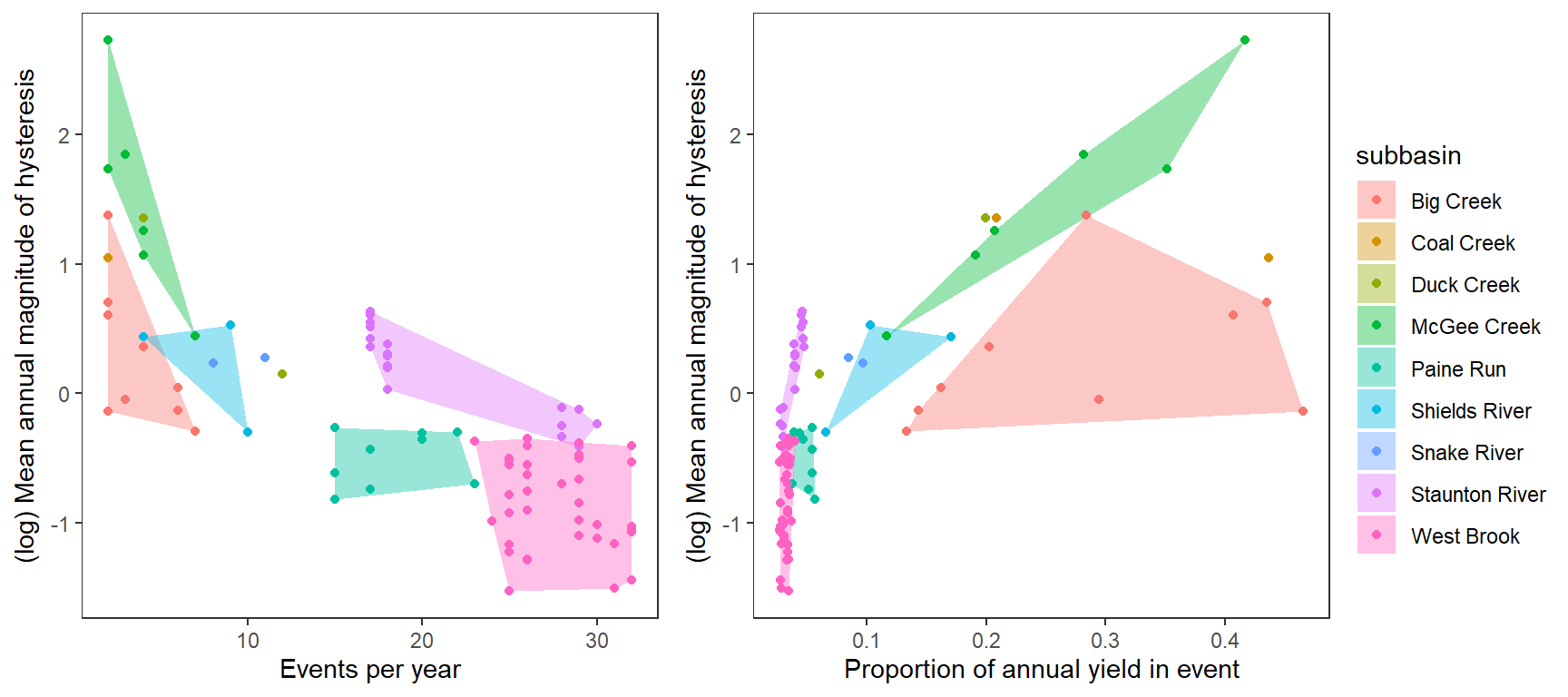
Alternatively, plot frequency by magnitude and with point color/size. I don’t think this tells the story as well as the convex hull plots.
p1 <- temp %>%
ggplot(aes(x = meanpropyield, y = freq)) +
# geom_polygon(data = hull, aes(fill = subbasin), alpha = 0.5) +
geom_point(aes(color = subbasin, size = logmeanhyst)) +
scale_size_continuous(range = c(0.1,5)) +
xlab("Proportion of annual yield in event") + ylab("Events per year") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
p2 <- temp %>%
ggplot(aes(x = meanpropyield, y = freq)) +
# geom_polygon(data = hull, aes(fill = subbasin), alpha = 0.5) +
geom_point(aes(color = logmeanhyst), size = 2) +
xlab("Proportion of annual yield in event") + ylab("Events per year") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
ggarrange(p1, p2, nrow = 1)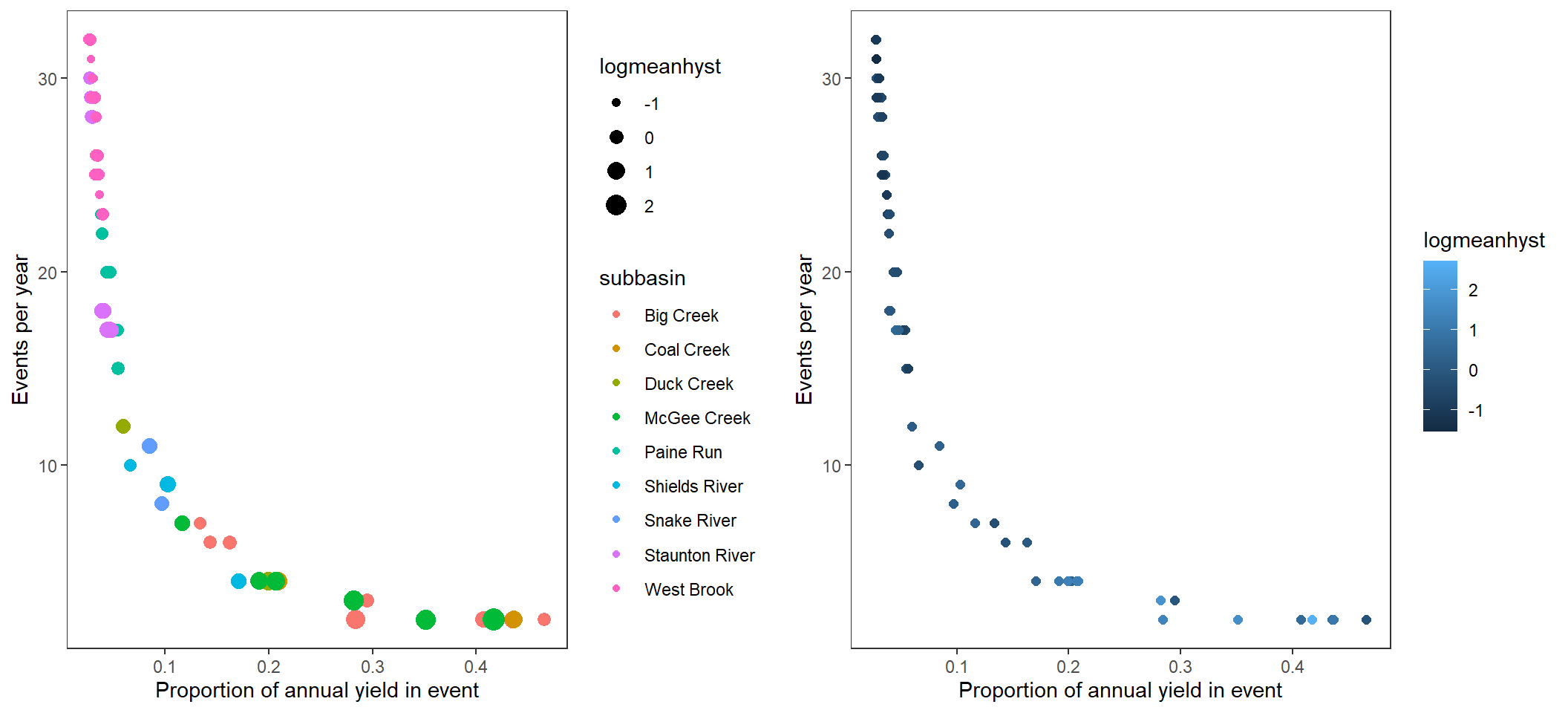
mycols <- c(brewer.pal(8, "Dark2"), "dodgerblue", "darkorchid")
events <- hysteresis %>% filter(subbasin == "Staunton River", WaterYear == 2020, eventid == 2)
length(unique(events$mindate))[1] 1length(unique(events$maxdate))[1] 1dat_clean_hyst %>%
filter(subbasin == unique(events$subbasin),
date >= unique(events$mindate),
date <= unique(events$maxdate)) %>%
ggplot(aes(x = logYield_big, y = logYield, group = site_name, color = site_name)) +
geom_segment(aes(xend = c(tail(logYield_big, n = -1), NA), yend = c(tail(logYield, n = -1), NA)),
arrow = arrow(length = unit(0.2, "cm")), color = "black") +
geom_point() +
geom_abline(intercept = 0, slope = 1, linetype = "dashed") +
#scale_color_gradientn(colors = cet_pal(250, name = "r1"), trans = "date") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())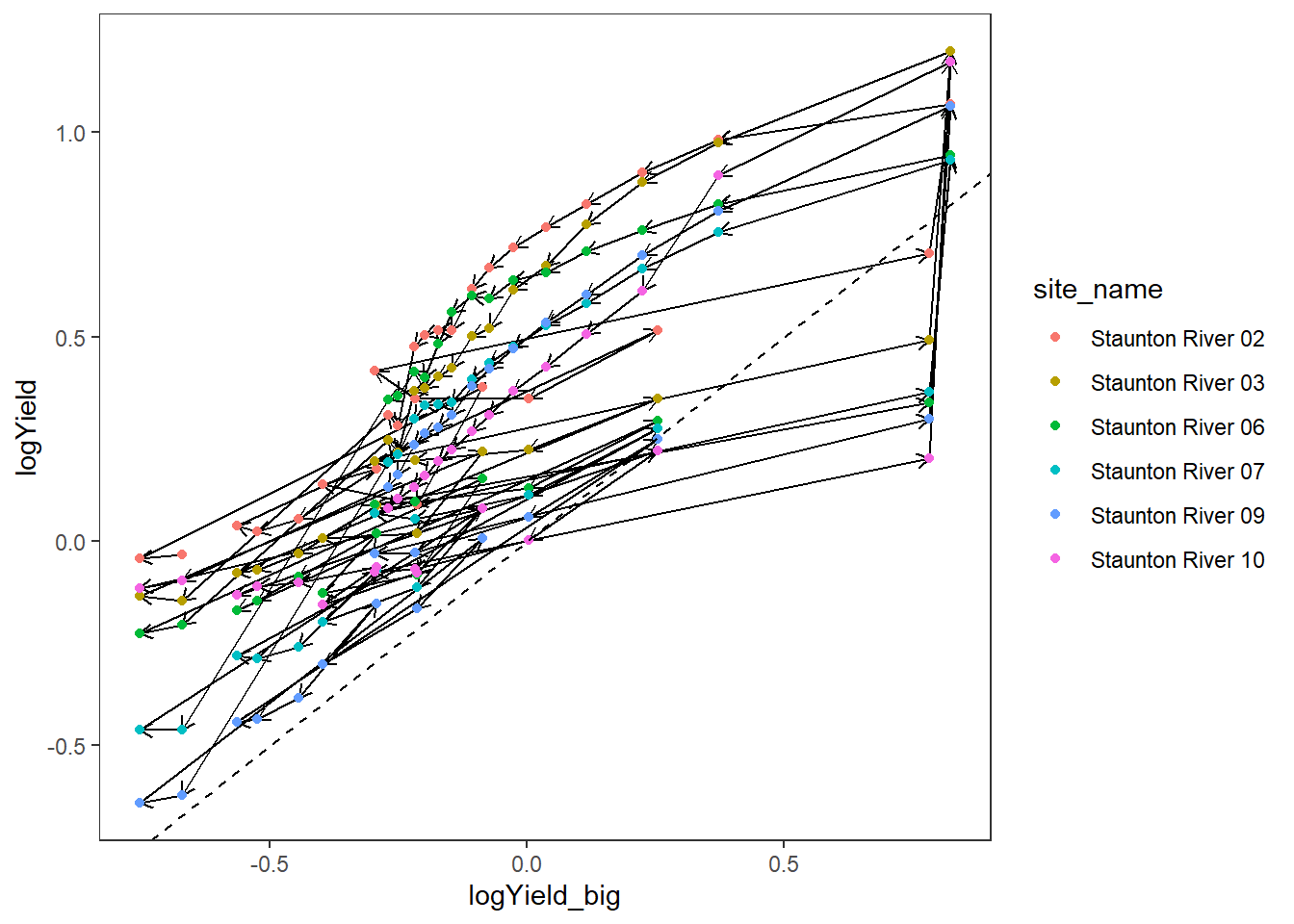
dat_clean_hyst %>%
filter(subbasin == unique(events$subbasin),
date >= unique(events$mindate),
date <= unique(events$maxdate)) %>%
ggplot(aes(x = logYield_big, y = logYield, group = site_name, color = site_name)) +
geom_segment(aes(xend = c(tail(logYield_big, n = -1), NA), yend = c(tail(logYield, n = -1), NA)),
arrow = arrow(length = unit(0.2, "cm")), color = "black") +
geom_point() +
geom_abline(intercept = 0, slope = 1, linetype = "dashed") +
#scale_color_gradientn(colors = cet_pal(250, name = "r1"), trans = "date") +
facet_wrap(~site_name) +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none")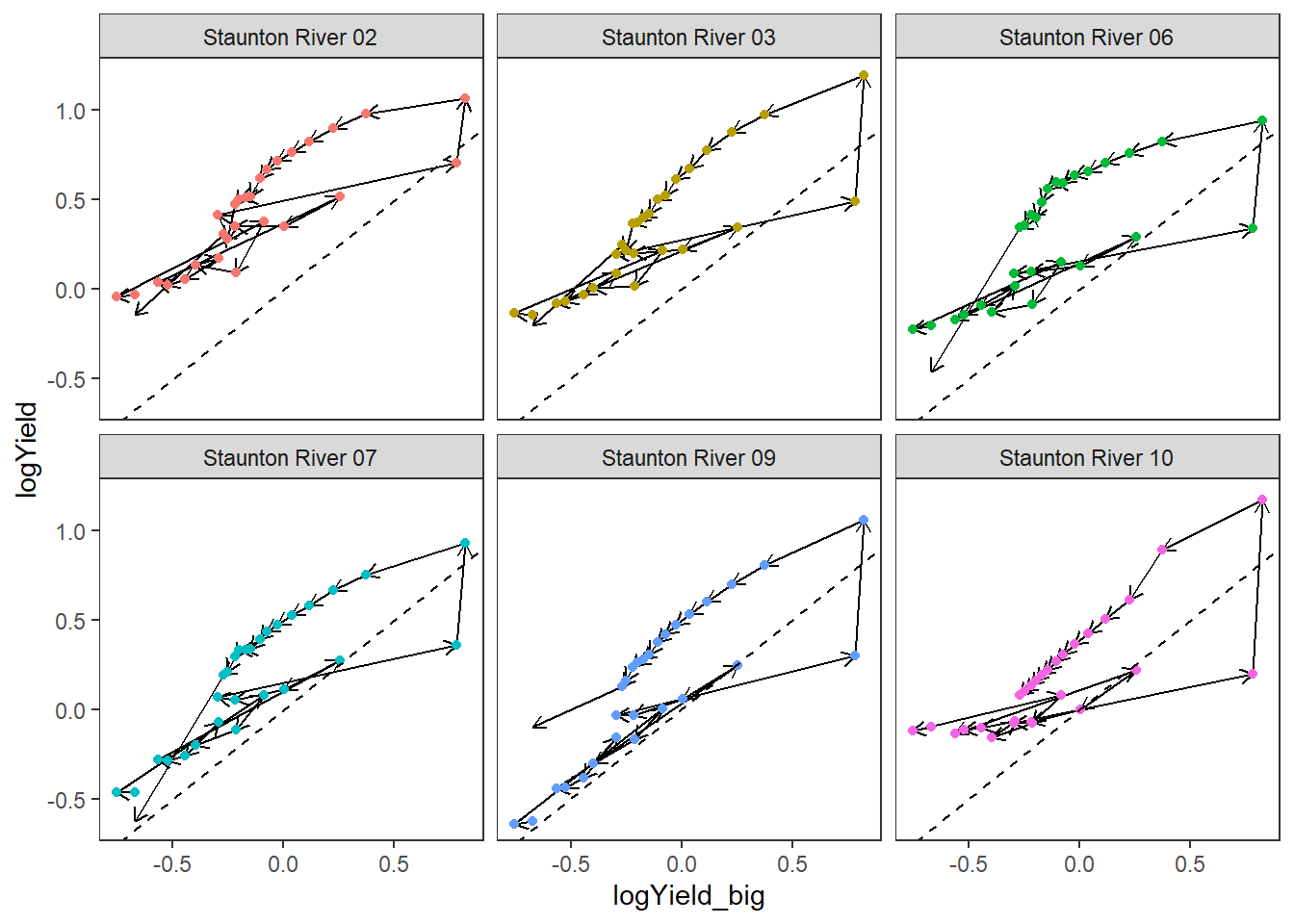
temp2 <- hysteresis %>%
#filter(subbasin == "West Brook") %>%
mutate(eventid_global = paste(WaterYear, eventid, sep = "-")) %>%
group_by(subbasin, eventid_global, mindate, totalyieldevent_big) %>%
summarize(nsites = n(), sdhyst = sd(hysteresis), meanhyst = mean(abs(hysteresis)), sd_totalyieldevent_little = sd(totalyieldevent_little)) %>%
ungroup()
temp2$antecedant <- NA
for (i in 1:dim(temp2)[1]) {
temp2$antecedant[i] <- sum(unlist(dat_clean_big %>% filter(subbasin == unique(temp2$subbasin), date < temp2$mindate[i], date >= temp2$mindate[i]-days(30)) %>% select(Yield_mm)))
}
head(temp2)# A tibble: 6 × 9
subbasin eventid_global mindate totalyieldevent_big nsites sdhyst meanhyst
<chr> <chr> <date> <dbl> <int> <dbl> <dbl>
1 Big Creek 2019-1 2018-10-26 14.9 3 0.815 2.08
2 Big Creek 2019-2 2019-03-21 188. 1 NA 4.91
3 Big Creek 2019-2 2019-03-21 291. 1 NA 2.18
4 Big Creek 2019-2 2019-03-21 352. 1 NA 1.77
5 Big Creek 2020-1 2019-12-21 2.38 3 0.355 0.937
6 Big Creek 2020-2 2020-01-31 2.19 2 0 0.226
# ℹ 2 more variables: sd_totalyieldevent_little <dbl>, antecedant <dbl>Larger events at big G (cumulative flow) tend to be associated with larger events at little g’s.
hysteresis %>% #filter(subbasin == "West Brook") %>%
mutate(eventid_global = paste(WaterYear, eventid, sep = "-")) %>%
ggplot(aes(x = totalyieldevent_big, y = totalyieldevent_little)) +
geom_point(aes(color = eventid_global)) +
geom_abline(intercept = 0, slope = 1, linetype = "dashed") +
geom_quantile(color = "black") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none") +
facet_wrap(~subbasin, scales = "free")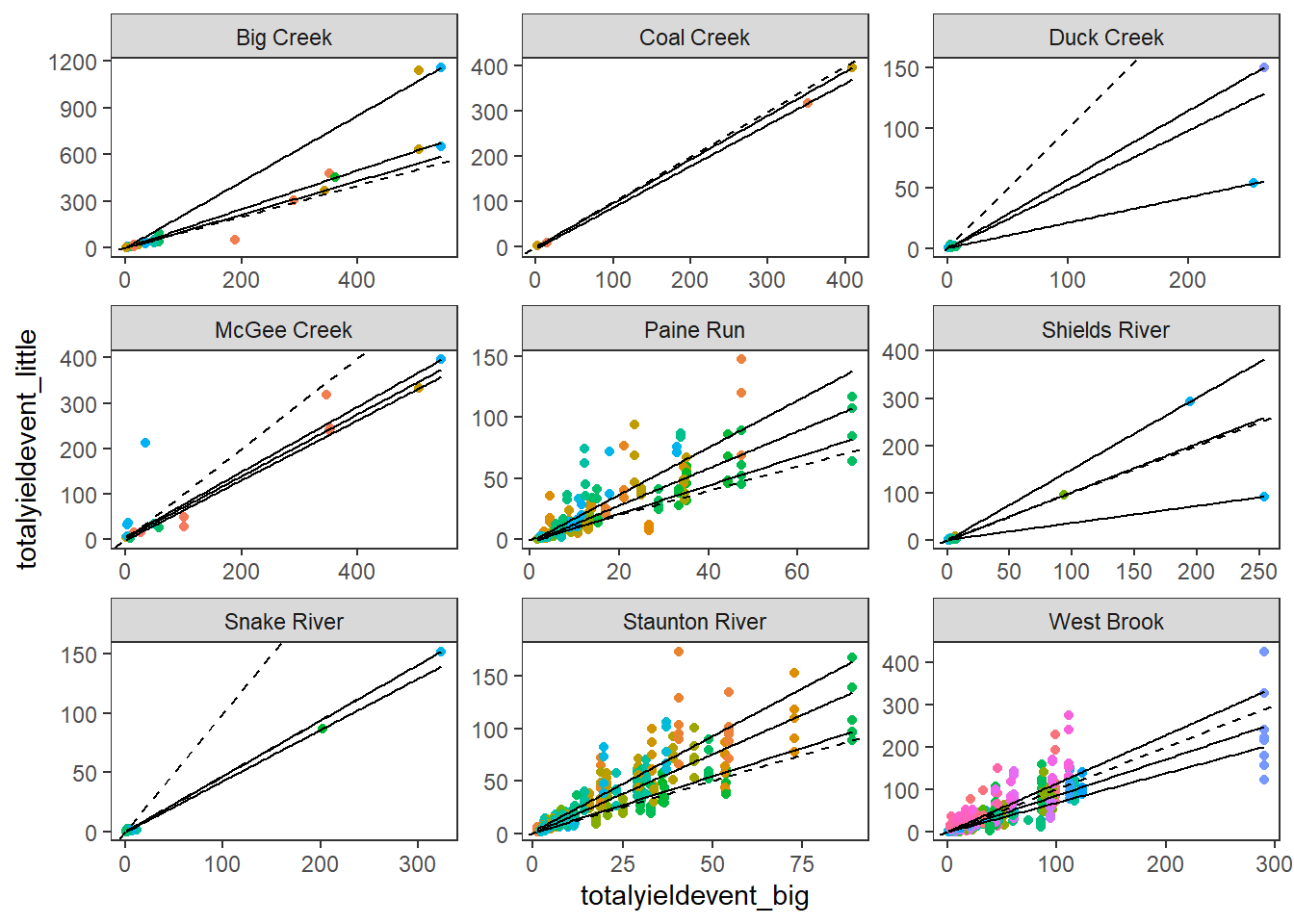
Variation among little g flow volume also increases with event magnitude.
temp2 %>%
ggplot(aes(x = log(totalyieldevent_big), y = log(sd_totalyieldevent_little))) +
geom_point() +
geom_smooth(method = "lm") +
xlab("(log) Cumulative yield at big G") + ylab("(log) SD cumulative yield at little G") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none") +
facet_wrap(~subbasin, scales = "free")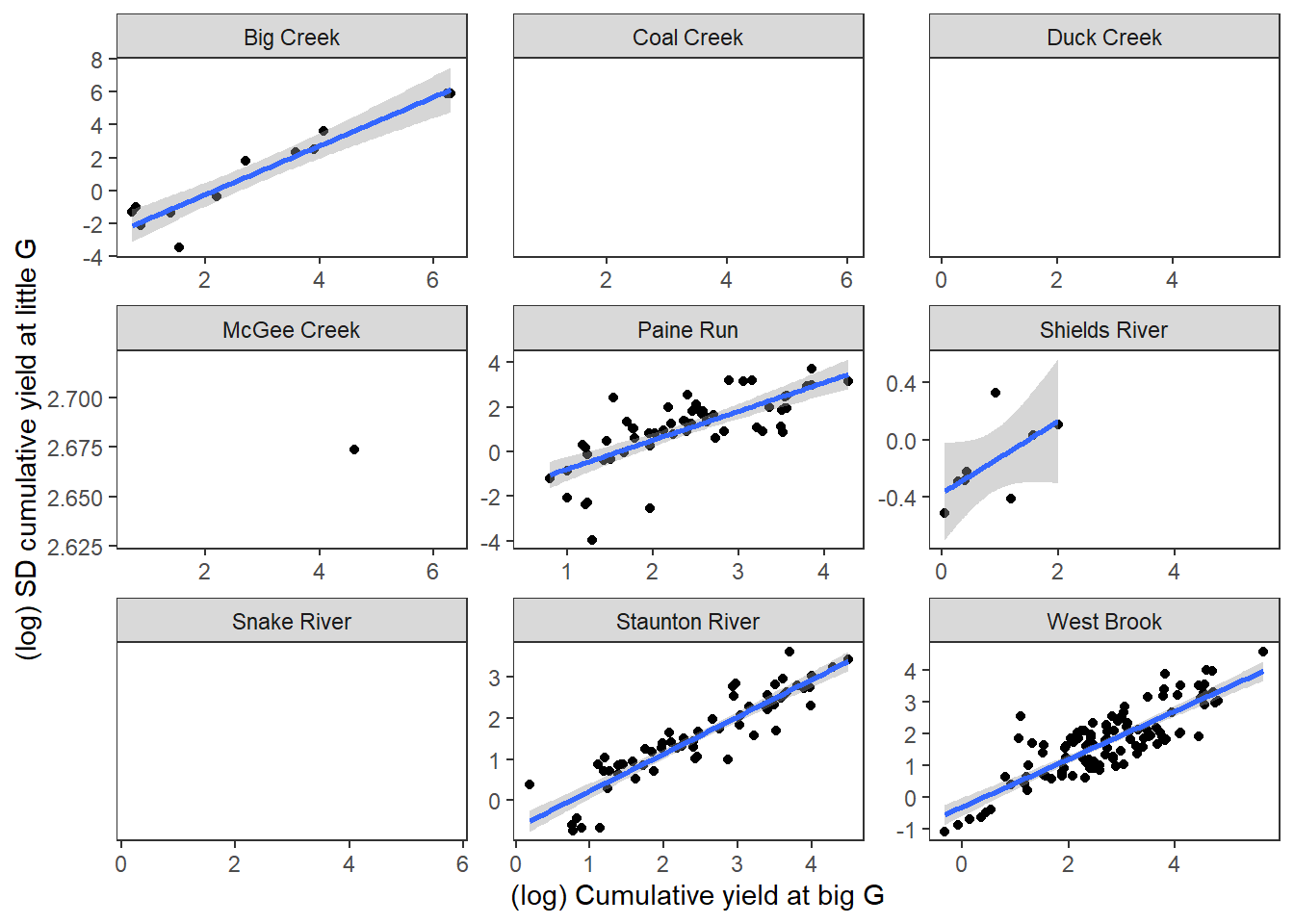
Events with greater average hysteresis also tend to be more variable among the little g’s
p1 <- temp2 %>%
ggplot(aes(x = meanhyst, y = sdhyst)) +
geom_point() +
geom_smooth(method = "lm") +
xlab("Mean hysteresis (among little g's)") + ylab("SD hysteresis (among little g's)") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none")
p2 <- temp2 %>%
ggplot(aes(x = log(meanhyst), y = log(sdhyst))) +
geom_point() +
geom_smooth(method = "lm") +
xlab("(log) Mean hysteresis (among little g's)") + ylab("(log) SD hysteresis (among little g's)") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none")
ggarrange(p1, p2, ncol = 2)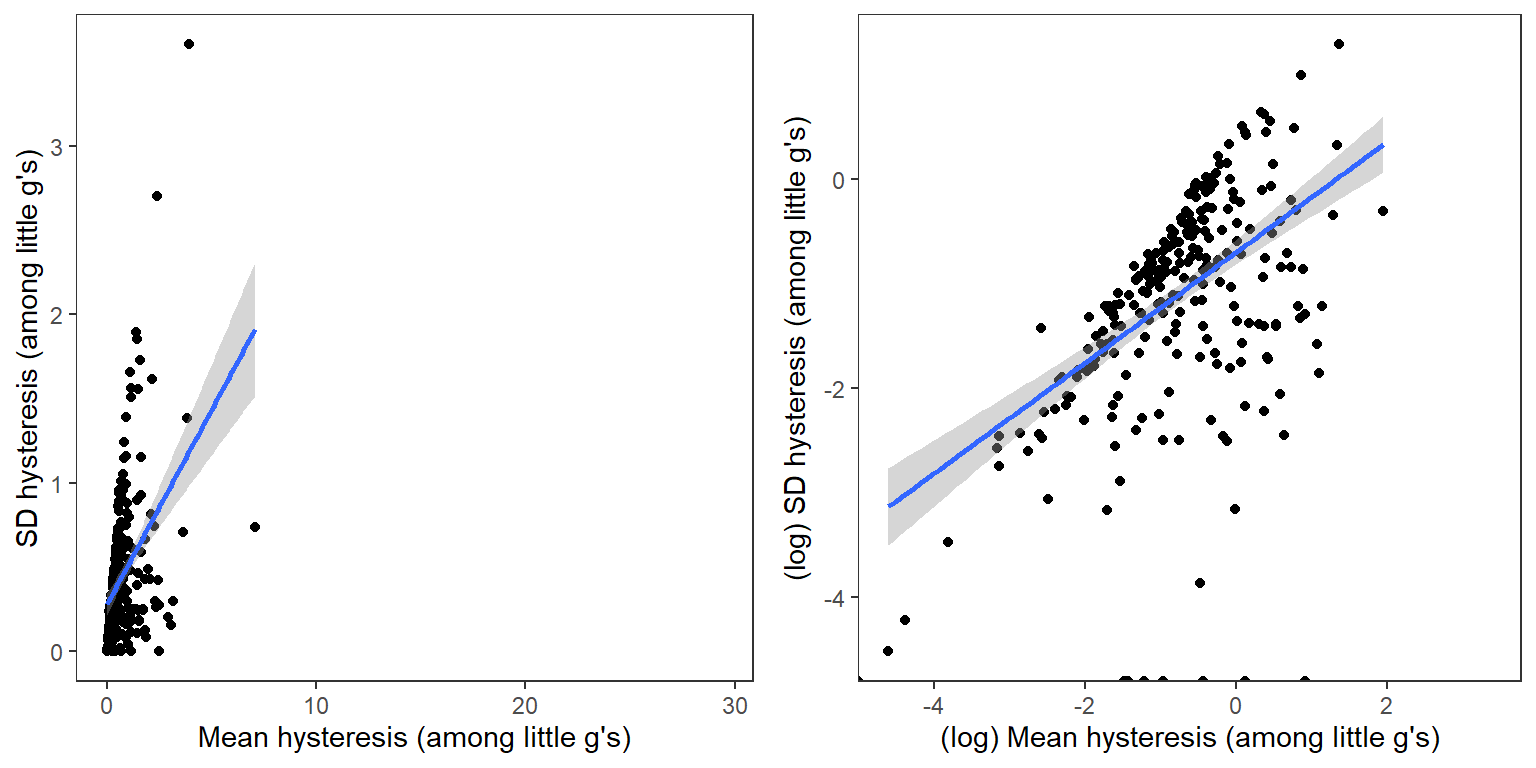
Generally, larger events (cumulative yield at big G) lead to greater mean hysteresis among little g’s and more variation in hysteresis among little g’s
p1 <- temp2 %>%
filter(sdhyst > 0) %>%
ggplot(aes(x = (totalyieldevent_big), y = (meanhyst))) +
geom_point() +
geom_smooth() +
xlab("Cumulative yield at big G") + ylab("Mean hysteresis") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none")
p2 <- temp2 %>%
filter(sdhyst > 0) %>%
ggplot(aes(x = (totalyieldevent_big), y = (sdhyst))) +
geom_point() +
geom_smooth() +
xlab("Cumulative yield at big G") + ylab("SD hysteresis)") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none")
p3 <- temp2 %>%
filter(sdhyst > 0) %>%
ggplot(aes(x = log(totalyieldevent_big), y = log(meanhyst))) +
geom_point() +
geom_smooth(method = "lm") +
xlab("(log) Cumulative yield at big G") + ylab("(log) Mean hysteresis") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none")
p4 <- temp2 %>%
filter(sdhyst > 0) %>%
ggplot(aes(x = log(totalyieldevent_big), y = log(sdhyst))) +
geom_point() +
geom_smooth(method = "lm") +
xlab("(log) Cumulative yield at big G") + ylab("(log) SD hysteresis") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none")
ggarrange(p1, p2, p3, p4, nrow = 2, ncol = 2)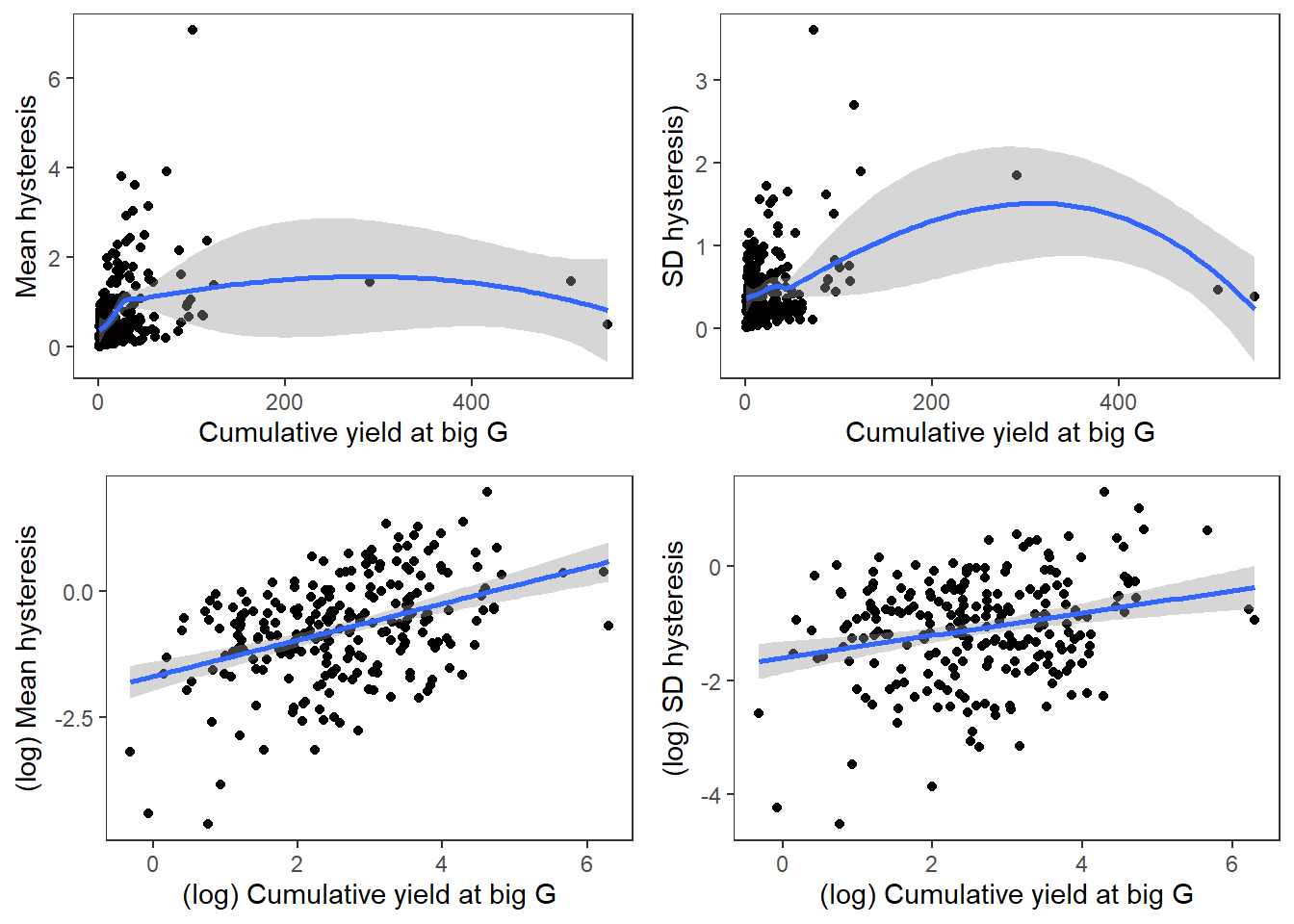
Events with greater average hysteresis also tend to be more variable among the little g’s
p1 <- temp2 %>%
ggplot(aes(x = antecedant, y = meanhyst)) +
geom_point() +
geom_smooth(method = "lm") +
xlab("Antecedant cumulative flow (30 days)") + ylab("Mean hysteresis") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none")
p2 <- temp2 %>%
ggplot(aes(x = antecedant, y = sdhyst)) +
geom_point() +
geom_smooth(method = "lm") +
xlab("Antecedant cumulative flow (30 days)") + ylab("SD hysteresis") +
theme_bw() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), legend.position = "none")
ggarrange(p1, p2, ncol = 2)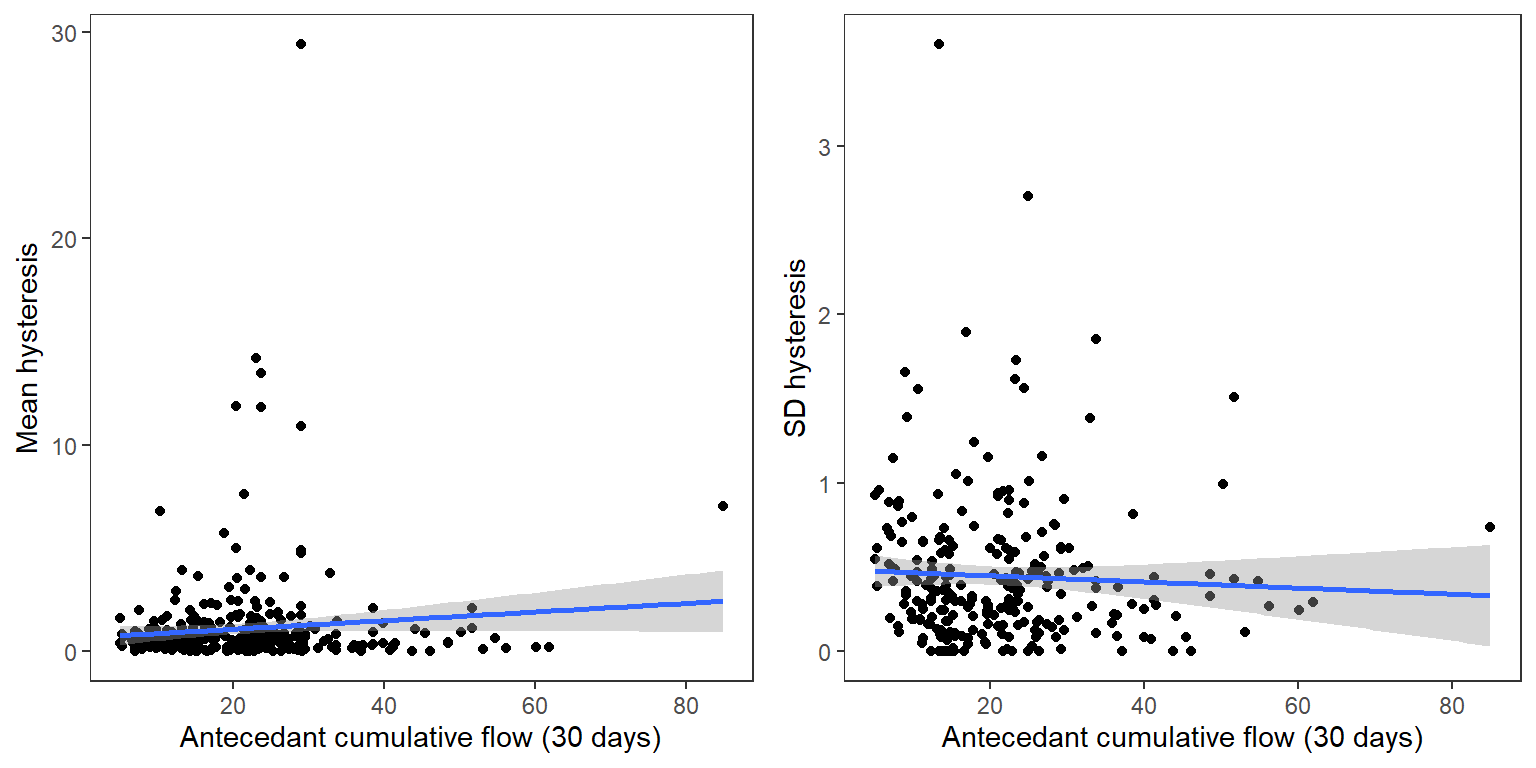
Conduct baseflow separation, event delineation, and dynamic time warping as above, but only for a single site year. For troubleshooting and data visualization
Set parameters and site-year
alp <- 0.95
numpass <- 3
thresh <- 0.75
dd <- dat_clean_hyst %>% filter(site_name == "McGeeCreekLower", WaterYear == 2019)baseflow separation
dd <- dd %>%
filter(!is.na(Yield_mm_big)) %>%
mutate(bf = baseflowB(Yield_mm_big, alpha = alp, passes = numpass)$bf,
bfi = baseflowB(Yield_mm_big, alpha = alp, passes = numpass)$bfi) %>%
ungroup()
head(dd)Event delineation
events <- eventBaseflow(dd$Yield_mm_big, BFI_Th = thresh, bfi = dd$bfi)
events <- events %>% mutate(len = end - srt + 1)
(events)Tidy events: now add variables to the Big G time series data specifying events and non-events
# define positions of non-events
srt <- c(1)
end <- c(events$srt[1]-1)
for (i in 2:(dim(events)[1])) {
srt[i] <- events$end[i-1]+1
end[i] <- events$srt[i]-1
}
nonevents <- data.frame(tibble(srt, end) %>% mutate(len = end - srt) %>% filter(len >= 0) %>% select(-len) %>% add_row(srt = events$end[dim(events)[1]]+1, end = dim(dd)[1]))
# create vectors of binary event/non-event and event IDs
isevent_vec <- rep(2, times = dim(dd)[1])
eventid_vec <- rep(NA, times = dim(dd)[1])
for (i in 1:dim(events)[1]) {
isevent_vec[c(events[i,1]:events[i,2])] <- 1
eventid_vec[c(events[i,1]:events[i,2])] <- i
}
# create vector of non-event IDs
noneventid_vec <- rep(NA, times = dim(dd)[1])
for (i in 1:dim(nonevents)[1]) { noneventid_vec[c(nonevents[i,1]:nonevents[i,2])] <- i }
# create vector of "agnostic events": combined hydro events and non-events
agnevents <- rbind(events %>% select(srt, end) %>% mutate(event = 1), nonevents %>% mutate(event = 0)) %>% arrange((srt))
agneventid_vec <- c()
for (i in 1:dim(agnevents)[1]){ agneventid_vec[c(agnevents[i,1]:agnevents[i,2])] <- i }
# add event/non-event vectors to Big G data
dd <- dd %>%
mutate(isevent = isevent_vec,
eventid = eventid_vec,
noneventid = noneventid_vec,
agneventid = agneventid_vec,
big_event_yield = ifelse(isevent_vec == 1, Yield_mm_big, NA),
big_nonevent_yield = ifelse(isevent_vec == 2, Yield_mm_big, NA),
big_event_quick = big_event_yield - bf) %>%
rename(big_bf = bf, big_bfi = bfi)
(dd)View time series data with Big G event delineation
dd %>% select(date, Yield_mm_big, Yield_mm, big_event_yield, big_nonevent_yield) %>% dygraph() %>% dyRangeSelector() %>% dyAxis("y", label = "Yield (mm)") %>% dyOptions(fillGraph = TRUE, drawGrid = FALSE, axisLineWidth = 1.5)View hysteresis loops: g~G
dd %>%
filter(isevent == 1) %>%
ggplot(aes(x = logYield_big, y = logYield, color = date)) +
geom_segment(aes(xend = c(tail(logYield_big, n = -1), NA), yend = c(tail(logYield, n = -1), NA)),
arrow = arrow(length = unit(0.2, "cm")), color = "black") +
geom_point() +
geom_abline(intercept = 0, slope = 1, linetype = "dashed") +
scale_color_gradientn(colors = cet_pal(250, name = "r1"), trans = "date") +
facet_wrap(~eventid) +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())Select and view specific event and normalize data
ddd <- dd %>% filter(eventid == 1, !is.na(logYield), !is.na(logYield_big))
dddd <- dd %>% filter(date %in% c(min(ddd$date)-1, max(ddd$date)+1))
ddd <- ddd %>%
bind_rows(dddd) %>%
arrange(date) %>%
mutate(weight = (logYield_big - min(logYield_big, na.rm = TRUE)) / (max(logYield_big, na.rm = TRUE) - min(logYield_big, na.rm = TRUE)),
yield_little_norm = (logYield - min(logYield, na.rm = TRUE)) / (max(logYield, na.rm = TRUE) - min(logYield, na.rm = TRUE)),
yield_big_norm = (logYield_big - min(logYield_big, na.rm = TRUE)) / (max(logYield_big, na.rm = TRUE) - min(logYield_big, na.rm = TRUE)))
ddd %>%
ggplot(aes(x = yield_big_norm, y = yield_little_norm, color = date)) +
geom_segment(aes(xend = c(tail(yield_big_norm, n = -1), NA), yend = c(tail(yield_little_norm, n = -1), NA)),
arrow = arrow(length = unit(0.2, "cm")), color = "black") +
geom_point() +
geom_abline(intercept = 0, slope = 1, linetype = "dashed") +
scale_color_gradientn(colors = cet_pal(250, name = "r1"), trans = "date") +
theme_bw() +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())Align data using dynamic time warping
align <- dtw(x = unlist(ddd %>% select(yield_big_norm)), y = unlist(ddd %>% select(yield_little_norm)), step = asymmetric, keep = TRUE)
# align <- dtw(x = unlist(ddd %>% select(Yield_mm_big)), y = unlist(ddd %>% select(Yield_mm)), step = asymmetric, keep = TRUE)
plot(align, type = "threeway")
plot(align, type = "twoway", offset = -1)Calculate hysteresis index as in Xue et al (2024)
ddd <- ddd %>% mutate(distance = align$index1 - align$index2)
ddd %>%
ggplot(aes(x = date, y = distance*weight)) +
geom_bar(stat = "identity") +
theme_bw()
sum(ddd$weight * ddd$distance) / sum(ddd$weight)